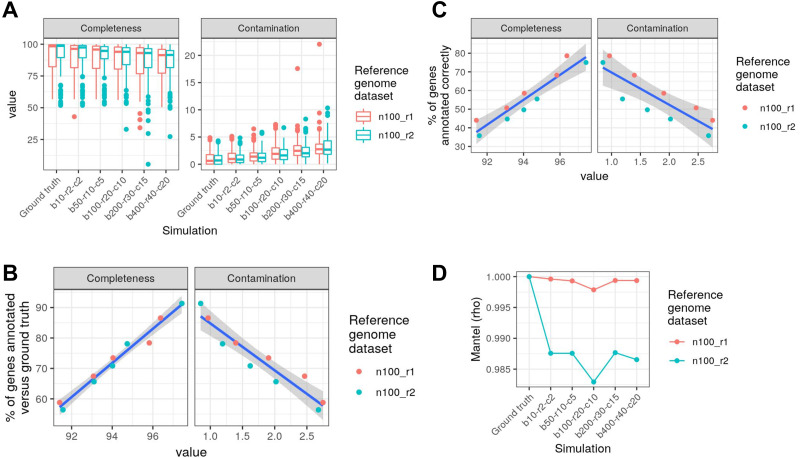
Figure 3. Struo2-generated gene database quality is substantially affected by reference genome assembly quality.
Two reference genome datasets of 100 randomly selected genomes each (“n100_r1” and “n100_r2”) were used for simulating misassemblies among all genomes in order to assess how genome assembly quality affects Struo2-generated database quality. “Ground truth” is the unaltered reference genomes, while the “bN-rN-cN” labels denote synthetic datasets with specific numbers of added misassemblies per genome (see Methods). (A) CheckM-estimated assembly quality for each genome. (B) The percent of genes annotated in the Struo2 database versus the ground truth. (C) The percent of genes annotated correctly (i.e., correct UniRef90 ID) versus the ground truth. (D) Change in Bray-Curtis distances between the ground truth and synthetic datasets (measured via Mantel tests), with beta diversity calculated from Bracken taxonomic assignments. The CAMI2 “HMP-gut” dataset of 10 metagenomes was used for benchmarking.