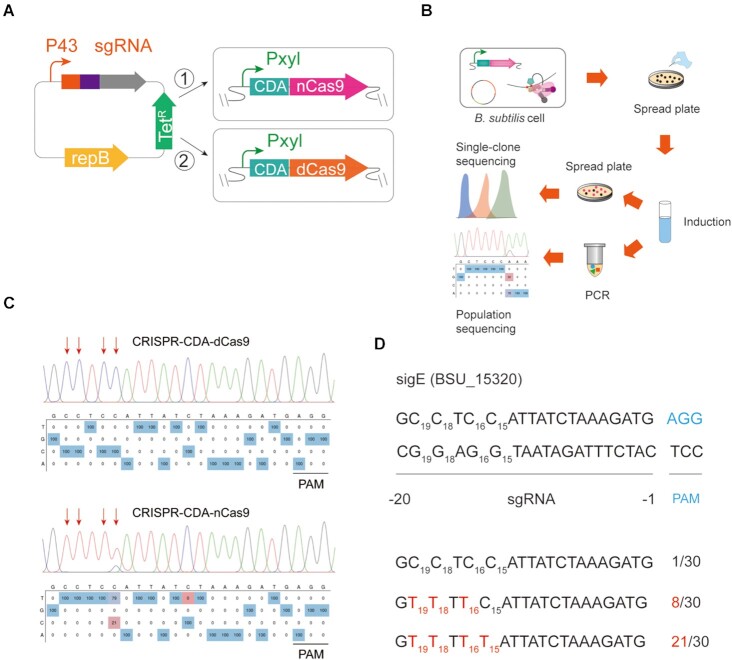
Figure 1.
Construction and characterization of CRISPR-CDA-Cas9-mediated base editor system in B. subtilis. (A) Design and construction of CRISPR-CDA-Cas9 mediating base editors. The fusion protein comprising nCas9/dCas9 and CDA under the control of xylose promoter PxylA was integrated into the lacA locus in B. subtilis. The repressor gene xylR was constitutively expressed in the expression cassette. A strong constitutive promoter P43 controlled the designed sgRNA. (B) Workflow for gene editing identification and verification at the single-clone level and population level in B. subtilis. (C) The editing efficiency of CRISPR-CDA-dCas9 and CRISPR-CDA-nCas9 at sigE site. (D) Base conversion targeting sigE at the single-clone level. Thirty colonies were selected and subjected to gene sequencing and the conversion efficiency was calculated. The PAM sequence is blue, whereas the modified bases are red.