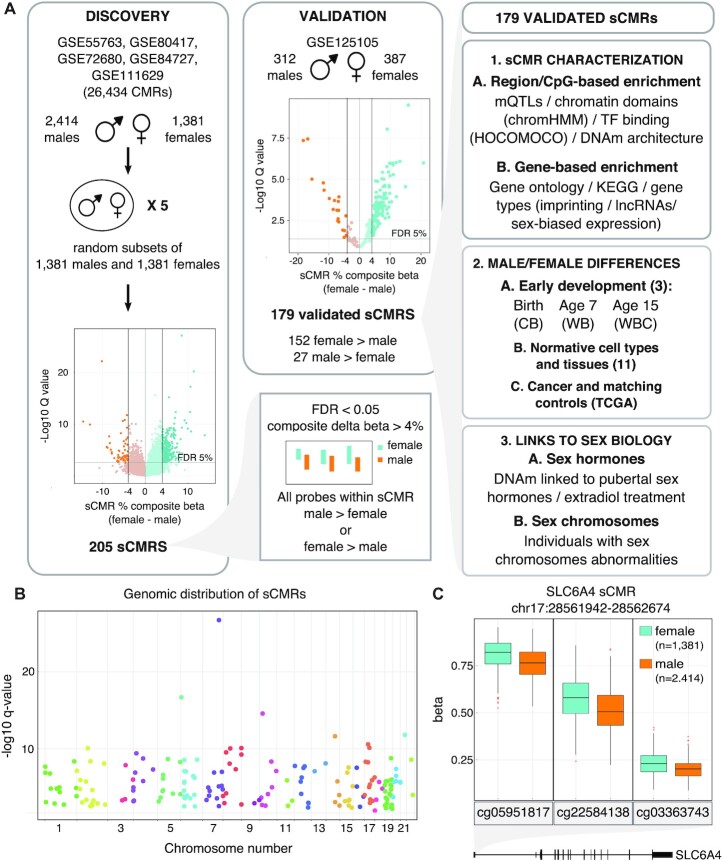
Figure 1.
sCMRs were reproducible, detected across the autosomes and included sites with known sex differences in DNAm levels. (A) Schematic of the sCMR discovery, validation and characterization steps. An aggregate cohort of 3795 (2414 males and 1381 females) adult whole blood DNAm samples was assembled and used to identify CMRs. Five sex-balanced random subsamples were generated and used to identify sCMRs. sCMRs were defined as CMRs having an FDR < 0.05% and a composite beta difference > 4% when comparing males and females. In addition, for a CMR to be considered all probes had to show the same sex-biased DNAm pattern, either higher DNAm levels in males compared to females or vice versa. (B) Manhattan plot showing the distribution of sCMRs across all the autosomes. (C) Boxplot showing male versus female DNAm levels for the three sites included in the SLC6A4 sCMR (chr17:28521337–28562986). cg05951817, cg22584138 and cg03363743 showed significant methylation (beta) differences of 6%, 8% and 3%, respectively, when comparing males and females (P-values of 0). The SLC6A4 sCMR was found at the 5′ UTR, a region previously shown to be differentially methylated between males and females in brain tissue (76).