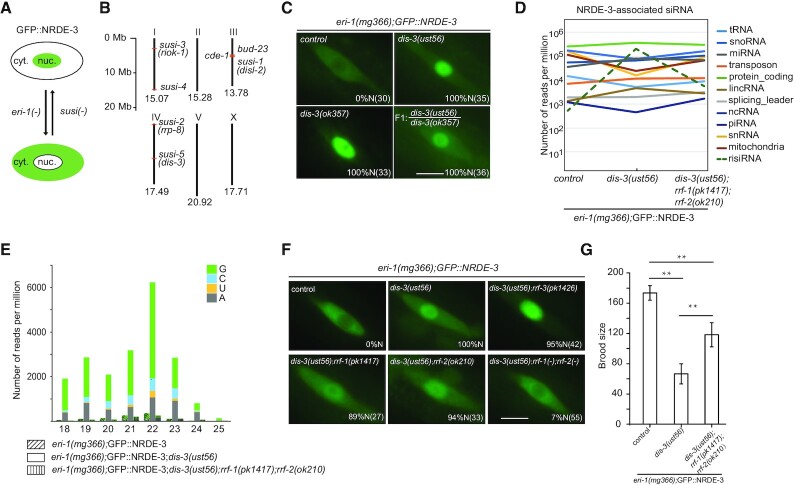
Figure 1.
A genetic screening identified the accumulation of antisense ribosomal siRNAs (risiRNAs) in dis-3 mutants. (A) The subcellular localization of NRDE-3 was used as an indicator to search for suppressors of endo-siRNA generation. cyt., cytoplasm; nuc., nucleus. (B) Summary of susi genes identified by forward genetic screening in C. elegans (5,14). Numbers indicate the size of each chromosome. (C) Images show seam cells of L4 stage animals expressing GFP::NRDE-3. Numbers indicate the percentage of animals with nucleus-enriched NRDE-3 in seam cells (% N). The number of scored animals is indicated in parentheses. Scale bars, 5 μm. (D) Results of the deep sequencing of NRDE-3-associated siRNAs from L3 stage animals. The green dashed lines indicate risiRNAs. The sense ribosomal reads and the sense protein-coding reads are discarded. (E) Size distribution and 5′-end nucleotide preference of NRDE-3-associated risiRNAs in L3 animals. (F) Images show seam cells of the respective L4 stage animals expressing GFP::NRDE-3, labeled as in (C). Scale bars, 5 μm. (G) Brood size of indicated animals grown at 20°C. Data are presented as the mean ± s.d.; n ≥ 15 animals; **P < 0.01.