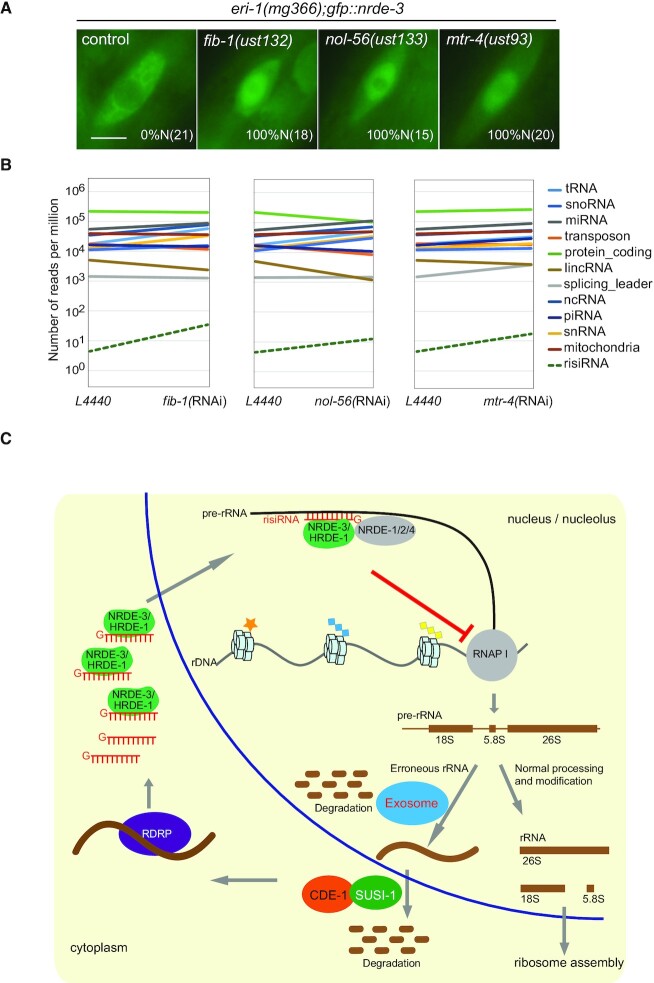
Figure 8.
risiRNAs were enriched in the mutants of rRNA processing and maturation factors. (A) Images of seam cells from L2 stage animals. Numbers indicate the percentages of the animals with nuclear-enriched NRDE-3 in seam cells (%N). The number of scored animals is indicated in parentheses. Scale bars, 5 μm. (B) Results from the deep sequencing of total small RNAs from the indicated animals. (C) A working model of risiRNA biogenesis and function. The processes of ribosome biogenesis are very sophisticated in eukaryotic cells from the splicing events of pre-rRNAs to the final assemblage of ribosomes, during which errors could occur at any step. In the nucleoli and nucleus, an exoribonucleolytic multisubunits protein complex, the exosome, participates in rRNA processing and intermediates degradation. In the cytoplasm, erroneous rRNAs are uridylated at the 3′-ends by polyuridylating polymerase-I (named CDE-1 or PUP-1) and then degraded by the 3′ to 5′ exoribonuclease DISL-2. Deficiency of these two degradation systems results in the accumulation of erroneous uridylated rRNAs, which further recruit additional RNA-dependent RNA polymerases (RdRPs) to synthesize risiRNAs and initiate the nucleolar gene silencing cascade. risiRNAs associate with the nuclear Argonaute protein NRDE-3 in soma or HRDE-1 in the germline, bind to pre-rRNAs, and inhibit RNAP I transcription elongation. Therefore, by combining the RNA degradation system with nucleolar gene silencing machinery, cells surveil the quality of rRNAs and maintain the rRNA homeostasis.