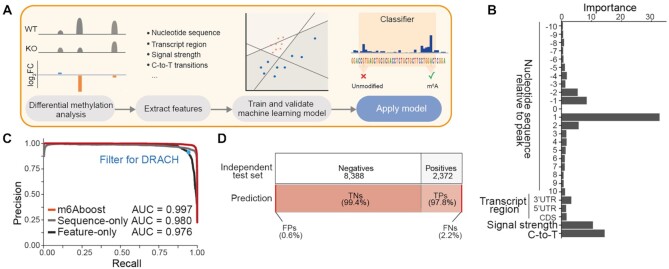
Figure 3.
The machine learning classifier m6Aboost reliably predicts m6A sites from miCLIP2 data. (A) Overview of the machine learning approach. First, miCLIP2 WT and Mettl3 KO datasets are analyzed for differential methylation to identify Mettl3-dependent m6A sites. The resulting positive and negative sets are used to extract features and train a machine learning classifier. The model is validated on an independent test set. Finally, the model can be applied to new miCLIP2 datasets to classify the miCLIP2 peaks as modified m6A sites versus unmodified background signal. (B) Highest informative content lies in the nucleotide sequence, the relative signal strength of the peak and the number of C-to-T transitions. Bar plot shows the features used for m6Aboost prediction and their associated importance ranking. UTR, untranslated region, CDS, coding sequence. (C) m6Aboost outperforms baseline models trained only on sequence (sequence-only) or experimental features (feature-only). Precision-recall curve shows performance of m6Aboost compared to baseline models with the corresponding area under the curve (AUC). Precision and recall when solely filtering for DRACH motifs are shown for comparison (blue dot). (D) m6Aboost achieves 99% accuracy on an independent test set. Bars visualize composition of independent test set (n = 10,760) from positive (22%) and negative (78%) peaks (top) and the resulting m6Aboost predictions (bottom). In total, 10,658 peaks (99%) were correctly predicted, while 102 peaks were misclassified. TNs, true negatives, TPs, true positives, FNs, false negatives, FPs, false positives.