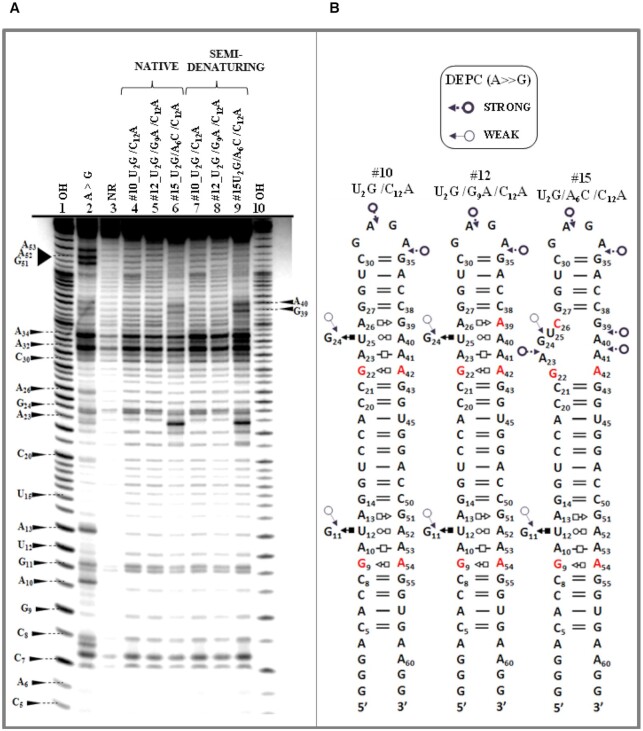
Figure 10.
Autoradiogram and summary of chemical probing analysis. (A) Structure probing using DEPC to probe accessibility of N-7 positions of Hoogsteen edges of A and G residues. Lanes: 1 and 10 are ‘OH−’: alkali hydrolysis step ladder of U2G/C12A hairpin, 2 is 'A > G'- Maxam-Gilbert sequencing reaction, 3 is non-reacted (‘NR’) U2G/C12A RNA, treated with aniline as control; NATIVE 4,5 and 6 are RNAs U2G/C12A, U2G/G9A/C12A, and U2G/A6C/C12A - samples treated with DEPC under native conditions (80mM NaCB buffer pH 6.94, 100mM KCl, 10 mM MgCl2, 0.5 mM EDTA); SEMI-DENATURING 7, 8 and 9 are RNAs U2G/C12A, U2G/G9A/C12A and U2G/A6C/C12A -samples treated with DEPC under semi-denaturing condition (80mM NaCB buffer pH 6.94, 100 mM KCl, 0.5 mM EDTA). (B) Summary of DEPC sensitivity given by legend on the top of the 2D structures.