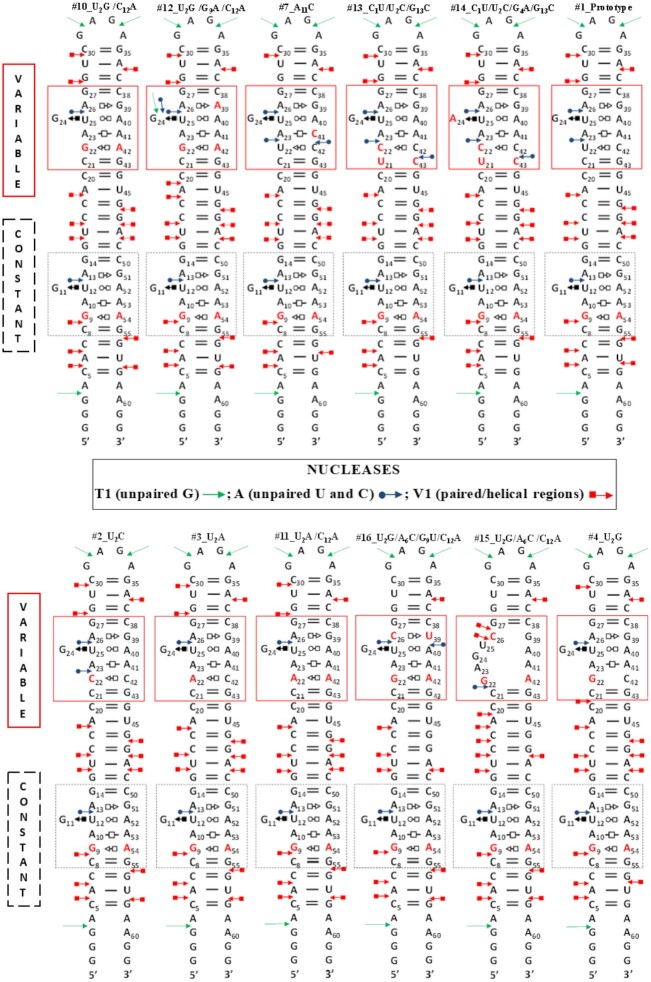
Figure 8.
2D diagrams of RNA hairpins used in structure probing experiments. These molecules consist of the same sequences of GAGA hairpin loop and have two distinct sequences regions: (i) constant region (highlighted as a puncture black rectangle) corresponds to sequence of molecule #10_U2G/C12A which conserved among all molecule and (ii) variable regions (highlighted as a red rectangle) implemented as a signature of individual molecule. The variable region contains sequences of duplexes used in thermal denaturation experiment. Probe RNA molecules are named according to the names of short duplexes inserted in variable region. For instance, prototype S/R motif has this name because the sequence of prototype S/R motif short duplex was implemented to the variable region. The summary of the nuclease digestion experiments is also shown. Cleavage patterns of RNA hairpin secondary structures is represented by symbols corresponding to the legend on center.