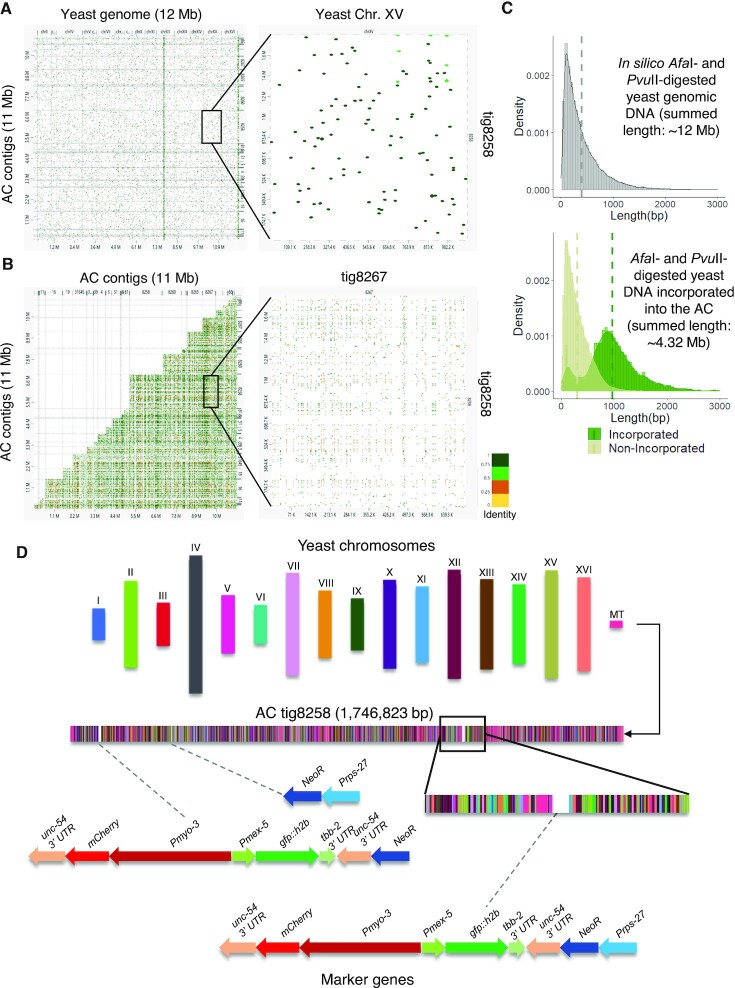
Figure 3.
De novo assembly results suggested that this AC is mostly formed by random non-homologous end-joining of the injected fragmented yeast genomic DNA and the NGM markers. (A) Whole genome alignment of assembled AC contigs to the yeast genome (Chromosome I to XVI) shows that the sequence of each contig is a random combination of short DNA fragments from different yeast chromosomes. The alignment between the largest contig tig8258 and yeast chromosome XV is magnified on the right. (B) Self-alignment of the contigs of the assembled AC shows that some short sequences are incorporated multiple times in different or the same contig. The alignment between the largest contig tig8528 and another contig tig8267 is magnified on the right. (C) Upper panel: a histogram plot of the DNA fragment lengths distribution density from in silico AfaI- and PvuII-digested budding yeast genome (summed-length 12 Mb). Bottom panel: histogram plots of the sequence length distribution density of sequences incorporated into the AC (summed-length ∼4.32 Mb, corresponding to larger fragments) and sequences that did not incorporate into the AC (summed-length ∼7.79 Mb, corresponding to smaller fragments). Dashed lines indicate the mean lengths. (D) A schematic representation of the distribution of yeast sequence fragments and the co-injection markers NGM (Prps-27::NeoR::unc-54 3′ UTR; Pmex5::gfp::tbb-2 3′ UTR; Pmyo-3::mCherry:: unc-54 3′ UTR) on the largest assembled contig tig8258. DNA fragments belong to individual yeast chromosomes and NGM marker genes are indicated by different colors. The full NGM marker sequences, or fragments inserted are shown in details below.