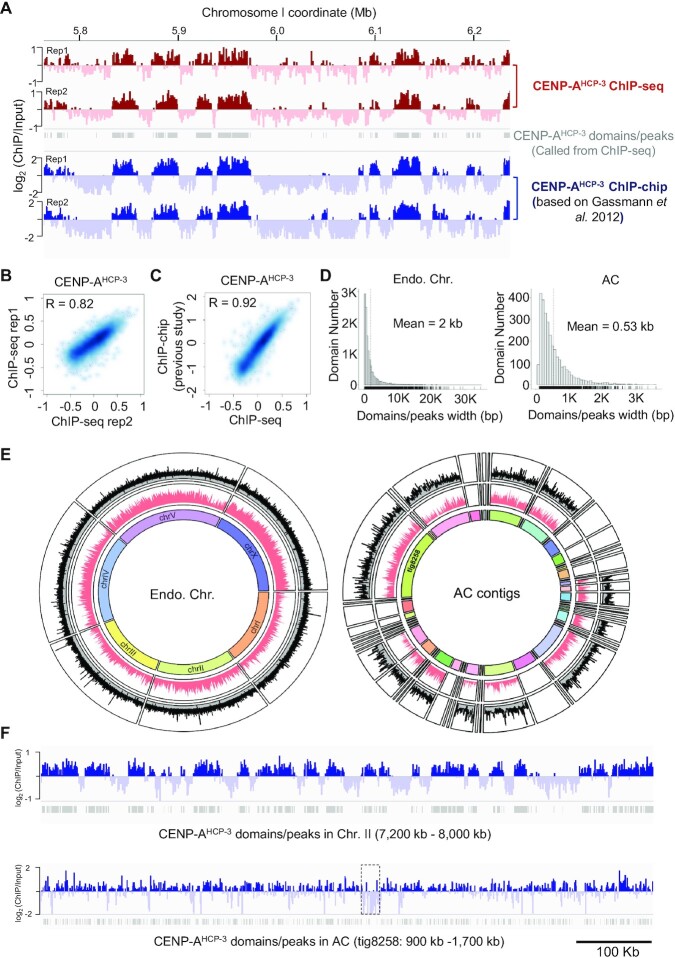
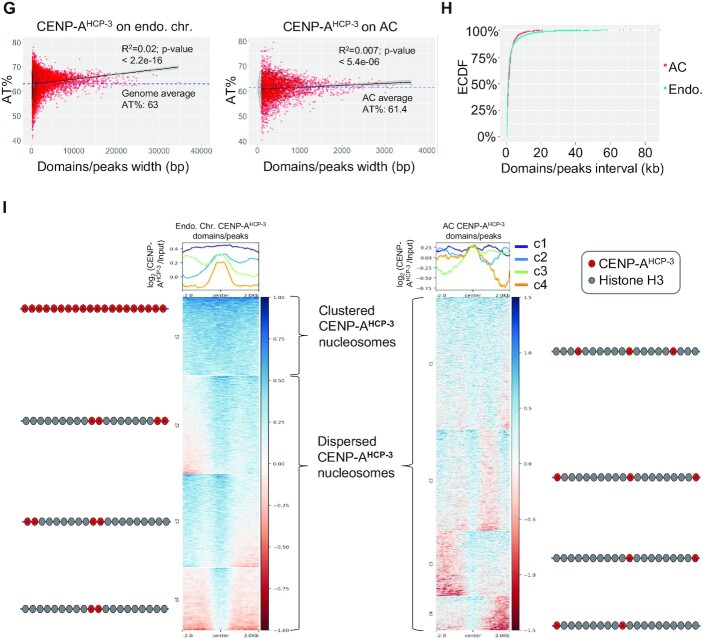
Figure 4.
The CENP-AHCP-3 domains/peaks on the endogenous chromosomes and on the prorogated AC. (A) Representative enrichment of CENP-AHCP-3 and CENP-AHCP-3 domains/peaks on chromosome I by current ChIP-seq replicates and previous ChIP-chip replicates. (B) The localization of CENP-AHCP-3 enrichment on endogenous chromosomes between two ChIP-seq replicates are highly correlated. (C) The localization of CENP-AHCP-3 enrichment on endogenous chromosomes detected by ChIP-seq is highly correlated to previous ChIP-chip results (18). The signal for each data set represents the average of two independent replicates. (D) Histogram plots of CENP-AHCP-3 domain/peak sizes in endogenous chromosomes (left panel) and in AC (right panel). (E) Circos plot of CENP-AHCP-3 domains/peaks on all endogenous chromosomes (left panel) and on all 49 AC contigs, including the longest contig tig8258 (right panel). The outside circle shows the CENP-AHCP-3 domains/peaks called by MACS2. Y-axis is the signal value (overall enrichment) for the domains/peaks. The inside circle indicate the CENP-AHCP-3 domains/peaks density in 10-kb windows. (F) Representative CENP-AHCP-3 enrichment in a 800-kb region in endogenous Chromosome II (7200–8000 kb) and in AC contig tig8258 (200–1000 kb). CENP-AHCP-3 domains/peaks were indicated as grey bars. The dashed box (1 319 126–1 347 939 bp) indicates the region enriched with yeast mitochondrial DNA (the zoom-in is shown in Figure 5A). (G) Dot plots of AT-content (%) of each CENP-AHCP-3 domain/peak against its size in endogenous chromosomes (left panel) and in AC (right panel). The 2D density plots were indicated by contour lines (bin = 7). The linear regression line to the scatter plot model was shown with 95% confidence region (grey shading). The average C. elegans genomic and AC AT-contents are indicated by the blue dotted lines, respectively. (H) The empirical cumulative distribution functions (ECDF) of interval distances between CENP-AHCP-3 domains/peaks on endogenous chromosomes (blue) and AC (red). (I) The CENP-AHCP-3 enrichment profiles and heatmaps of the 2-kb flanking regions surrounding the center of each CENP-AHCP-3 domain/peak for endogenous chromosomes and AC. Consensus CENP-AHCP-3 domains/peaks are separated into four clusters by k-means clustering. The profile plots (upper panel) show the average ChIP signals of each cluster. The ChIP signals of each domain/peak are shown in the clustered heatmaps (lower panel), in which the color coding on the right indicates the log2 CENP-AHCP-3 ratio. The schematic on the left and right show the possible distribution of CENP-AHCP-3 nucleosomes interspaced in between H3 nucleosomes.