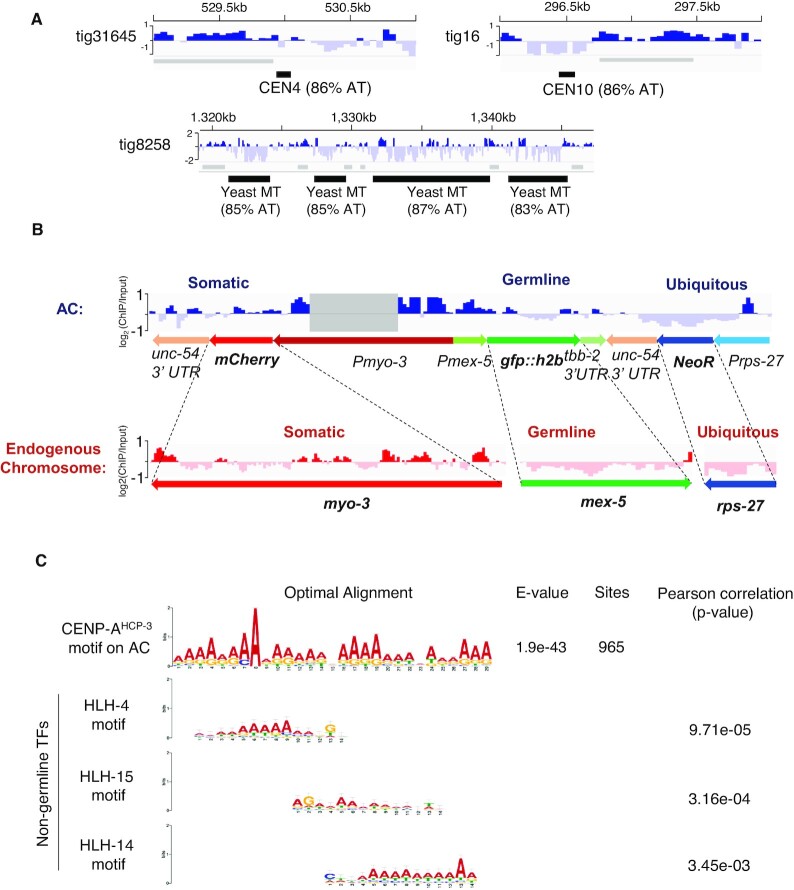
Figure 5.
The CENP-AHCP-3 distribution pattern on the propagated AC. (A) CENP-AHCP-3 does not localize on the yeast centromeric sequences, CEN4 and CEN10, and yeast mitochondrial DNA (MT) (the dashed box in Figure 4F) in the propagated AC. CENP-AHCP-3 domains were indicated as grey bars. (B) The log2 CENP-AHCP-3 ratio on marker genes is averaged from 10–29 copies (Supplementary Table S9). CENP-AHCP-3 on the AC is enriched in the somatic mCherry marker, partially excluded in the silenced germline gfp::h2b region, and excluded in the ubiquitous NeoR drug resistance gene marker. The corresponding endogenous genes with the same promoters were analyzed and shown in parallel. Non-distinguishable reads that mapped to both endogenous and AC promoter or 3′ UTR regions were excluded from the analysis and the region with all reads removed was shaded. (C) The sequence of a 29-bp CENP-AHCP-3 motif: AAAARRAARARAADVAAAAAAARARRAAA, is identified at 965 sites on the AC, where R represents A or G; D represents A or G or T; V represents A or C or G. The e-value estimates the expected occurrence of motifs with the same size and frequency in a similarly sized set of random sequences. Toggle error bars indicate the confidence level of a motif based on the number of sites used in its creation. The motifs found in the UniPROBE database with a significant match to the CENP-AHCP-3 motif on the AC are shown. The P-value indicates the significance of the similarity between CENP-AHCP-3 motif and TF motif.