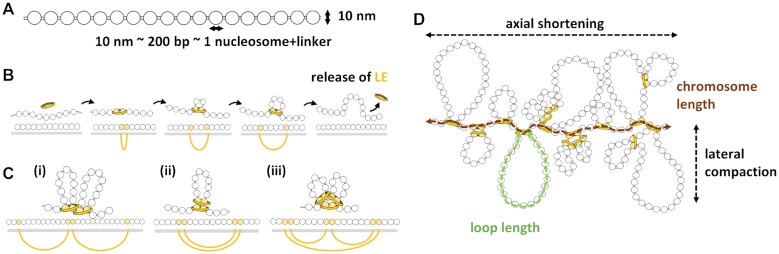
Figure 2.
Schematic representation of the adopted chromosome and loop extrusion models. (A) The chromosomal 10 nm chromatin fibre represented as a beads-on-a-string polymer. Around ∼200 base pairs of DNA (including the linker) are wrapped around each nucleosome. (B) The loop extrusion model. The Loop Extruder (LE) is represented by a yellow ring. Nucleosomes bound by LEs are shown in yellow, the bond between them is represented as a yellow ellipsoid line, and the grey bar represents the chromatin. (C) Different examples of loops formed by two proximal LEs: (i) side-by-side loops, (ii) nested loops and (iii) combination of both. (D) Chromosome condensation by loop formation. The bases of the loops form the axis of the chromosome, and the loops are radially distributed. The degree of chromosome condensation is due to axial shortening and lateral compaction. These two parameters are functions of the number of nucleosomes and can be computed as the chromosome length and the average loop length, respectively. In this example, the chromosome length is 18 axial nucleosomes and the loop length is 23 nucleosomes. We define axial nucleosomes as bound by LEs and outside any loop.