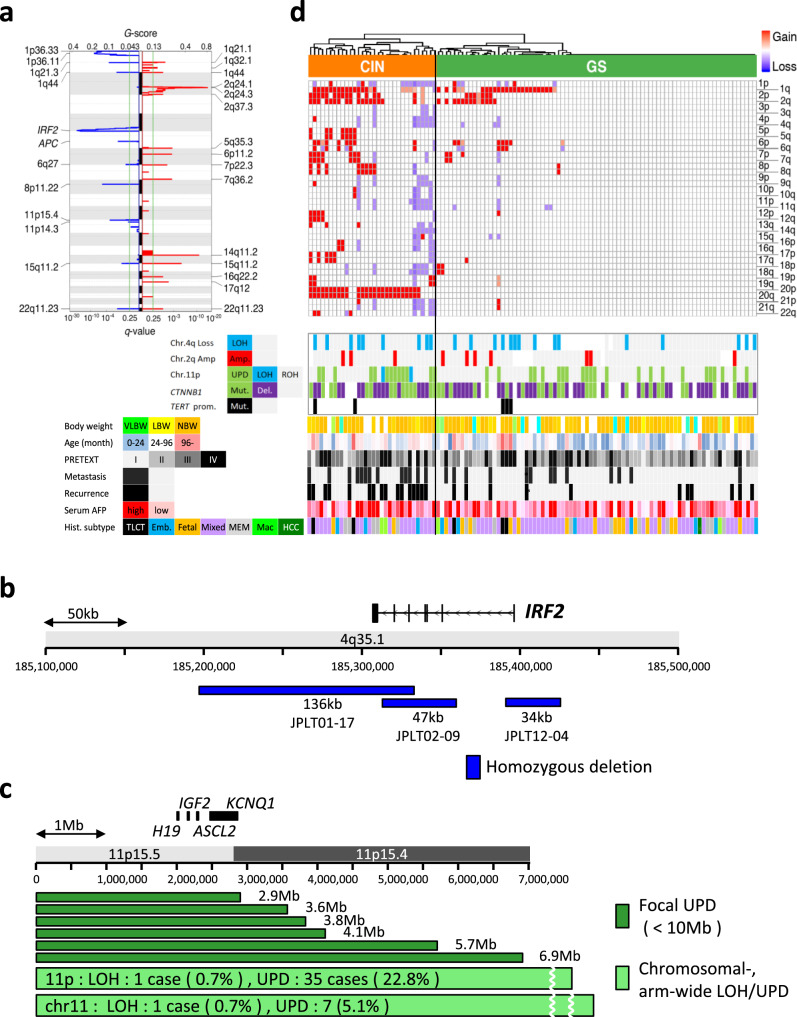
Fig. 2. Copy number aberrations and genomic stability of childhood hepatoblastoma (HB) genomes.
a Arm-wide chromosomal gain (left) and loss (right) were analyzed across 112 HB genomes using GISTIC 2.0. b Epicenter mapping of recurrent homozygous deletions in the vicinity of IRF2 gene detected in 3 HB cases. c Epicenter mapping of focal LOH and UPD in chromosome 11p spanning H19, IGF2, ASCL2, and KCNQ1 genes. d, A total of 112 HB tumors are hierarchically clustered into two groups based on arm-level somatic copy number alterations (SCNAs) defined by GISTIC 2.0; CIN (chromosomal instability, n = 32) and GS (genomically stable, n = 80). The heat map shows copy number statuses in each tumor (horizontal axis) plotted by chromosomal location (vertical axis) in the top panel. The middle panel represents the status of focal copy number (CN) changes in chromosomes 2q and 4p, LOH or UPD in chromosome 11p, and the driver mutations (CTNNB1, TERT). The bottom of the panel shows clinical and pathological parameters of HB patients. Statistical significance was analyzed by Fisher’s exact test (*p < 0.01).