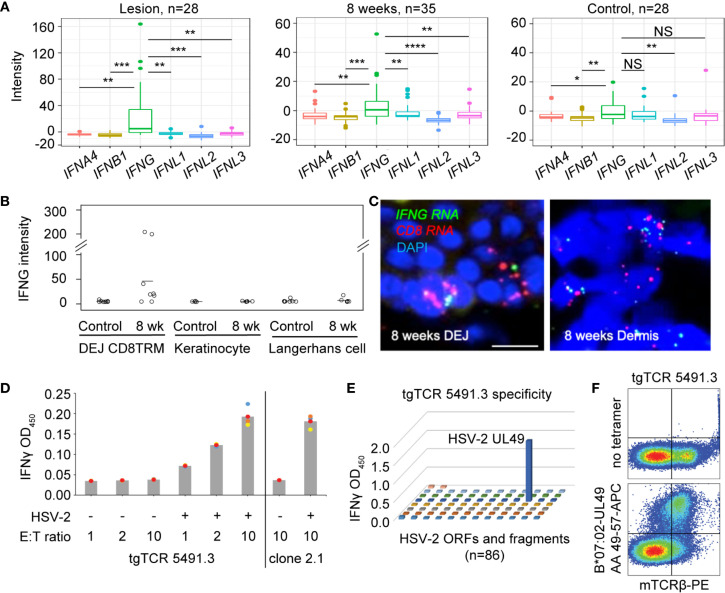
Figure 3.
Expression of IFNG predominates in HSV-affected skin during and after viral recurrence. (A) Transcriptional analysis showing expression levels of IFN4A, IFNB1 (type I IFN), IFNG (type II IFN), IFNL1, IFNL2 and IFNL3 (type III IFN) gene in skin biopsies of n = 28 at the time of HSV-2 lesion, n = 35 at the time of 8-week post-healing, and n = 28 matched contralateral normal controls. In box plots, the center line is the median, box edges show upper and lower quartiles, whiskers represent the range and dots indicate outliers. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS, not significant; student t test. (B) IFNG gene expression in DEJ CD8TRM (n = 8), keratinocytes (n = 5) and Langerhans cells (n = 7) laser microdissected from skin biopsies obtained at 8-week post-healing and their matched normal control skin, profiled by whole genome array. (C) RNA FISH images depicting co-expression of IFNG and CD8 mRNA transcript in cells (nuclei stained with DAPI) at the DEJ and in the dermis of an 8 weeks post-healing skin biopsy. Scale bars, 20 µm. (D) Recognition of autologous HSV-2-infected EBV-LCL by reconstructed transgenic TCR (tgTCR) 5491.3 reporter cells. Effectors and APC were co-incubated at indicated ratios for 24 hours. T cell activation as detected by IFN-γ release. HSV-2-specific CD8 T cell clone 2.1 as positive control. All combinations of effector cells and APC were run in quadruplicate. Each dot an individual value and bars as mean values. (E) Representative raw data from HSV-2 proteome-wide screen. Cos-7 cells were co-transfected with HLA B*07:02 cDNA and 86 individual HSV-2 genes or fragments. tgTCR5491.3 reporter cells were added for 24 h and IFN-γ release measured by ELISA. (F) tgTCR5491.3 reporter cells stained with anti-mouse TCR beta and tetrameric HLA B*07:02-HSV-2 UL49 AA 49-57 or no tetramer.