Fig. 2.
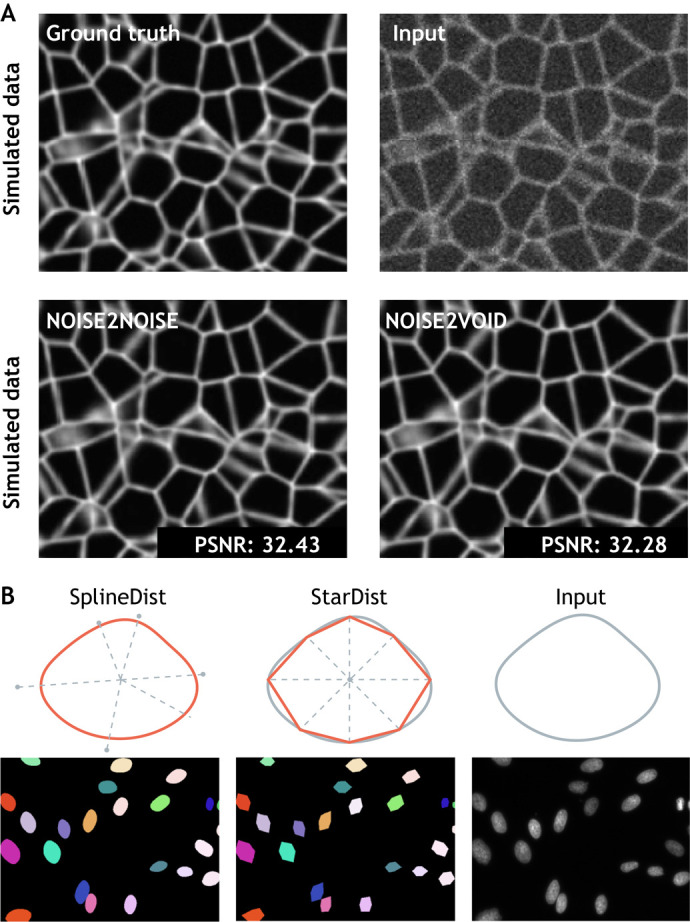
Deep learning methods applied to developmental biology applications. (A) A simulated ground truth cell membrane image is artificially degraded with noise. Denoised outputs obtained using Noise2Noise and Noise2Void are shown at the bottom, along with their average peak signal-to-noise values (PSNR; higher values translate to sharper, less-noisy images). Image adapted from Krull et al. (2019). (B) Fluorescence microscopy cell nuclei image from the Kaggle 2018 Data Science Bowl (dataset: BBBC038v1; Caicedo et al., 2019) segmented with StarDist (Schmidt et al., 2018), in which objects are represented as star-convex polygons, and with SplineDist, in which objects are described as a planar spline curve. Image adapted from Mandal and Uhlmann (2021).
