Fig. 1 |. Overview of spatial proteomics approaches.
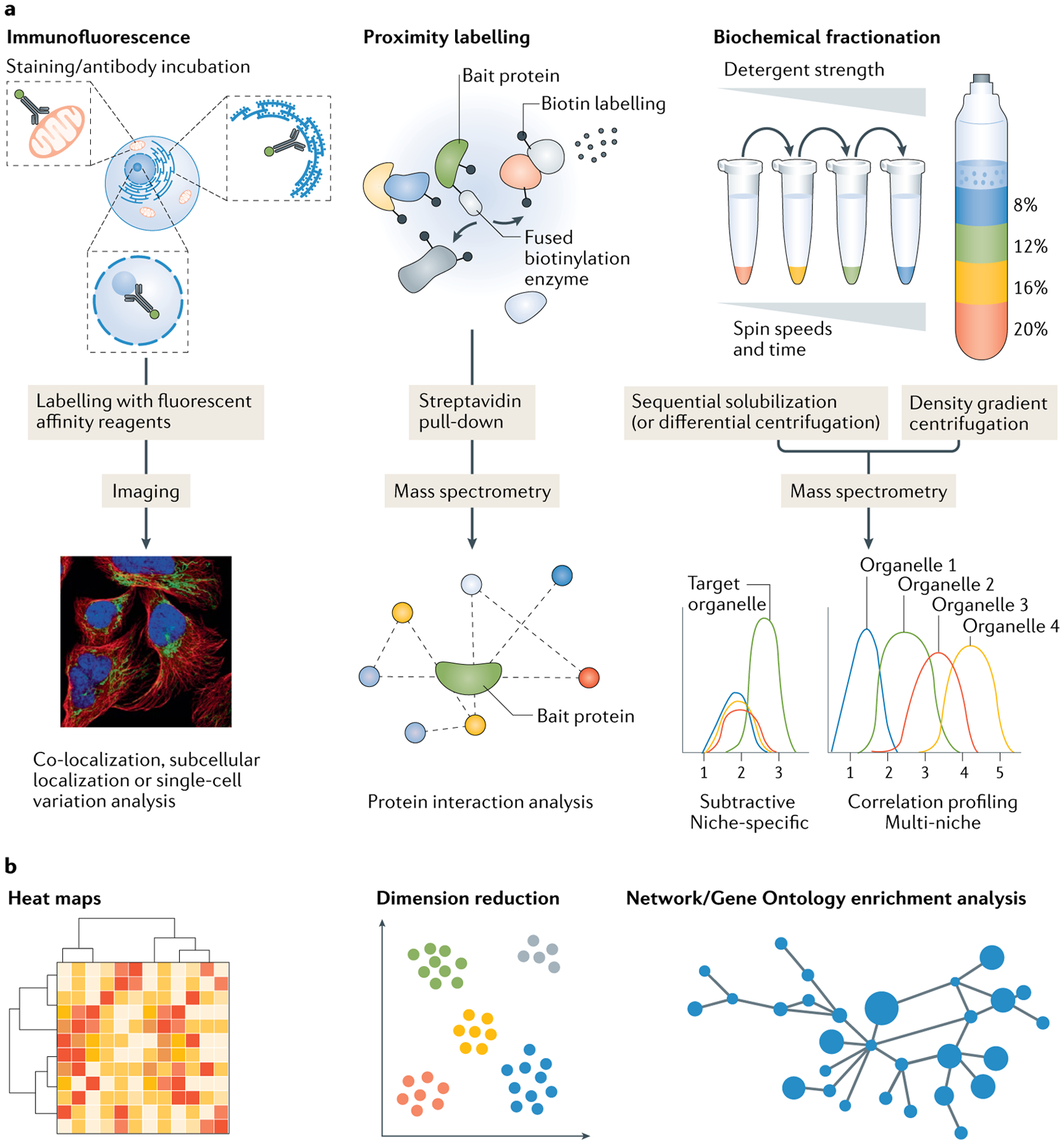
Spatial proteomics approaches include fluorescence imaging approaches and proximity labelling or biochemical fractionation techniques coupled with mass spectrometry (MS). a | Imaging of cells and tissues stained with fluorescently labelled antibodies (or other affinity reagents) allows for subcellular protein localization in situ. Proximity labelling strategies permit in vivo biotin labelling of proteins in close proximity to a chosen bait protein that has been genetically fused to a biotinylating enzyme. Following labelling, samples can be processed using MS proteomics protocols. Biochemical fractionation methods can produce cell fractions that are enriched for organelles of interest based on the different biophysical and chemical properties of different subcellular niches. These fractions are then subject to MS analysis. Typically, organellar separation is achieved using density gradient or differential/sedimentation centrifugation, or sequential solubilization using detergents190,191,303–305,315. b | All of these methods produce data-rich outputs that require computational analysis using techniques such as hierarchical clustering, dimension reduction or network analysis to visually represent and calculate statistical information. Machine learning techniques can also be used (not pictured). Correlation profiling plot in part a and dimension reduction plot in part b adapted from REF.21, Springer Nature Limited.
