Fig. 2 |. Proximity labelling proteomics.
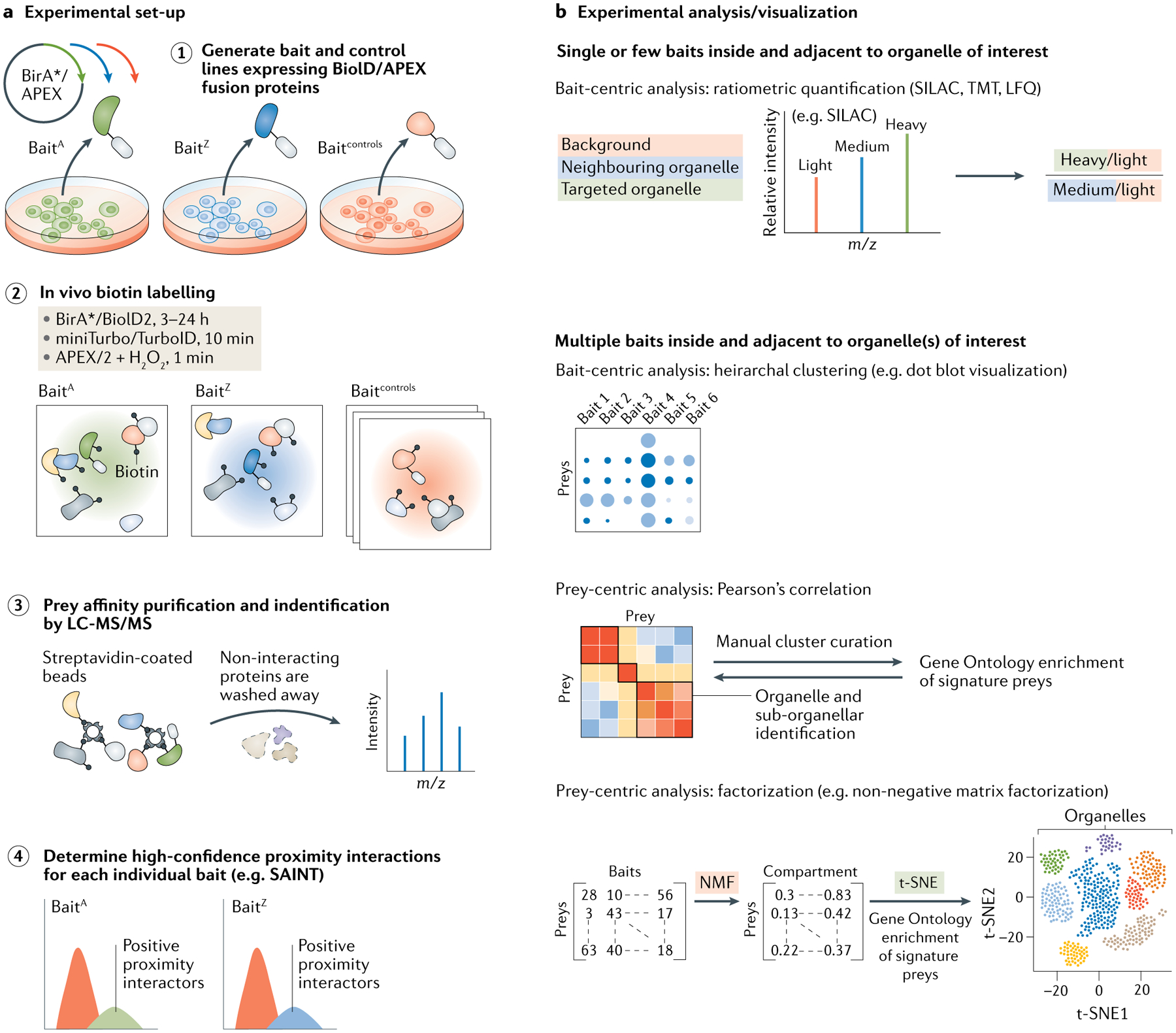
Proximity labelling strategies permit biotin labelling of proteins in immediate proximity to the chosen bait proteins in living cells. a | Baits of interest (BaitA, BaitZ) are genetically fused with an enzyme such as APEX/APEX2 or BirA*, BioID2, miniTurbo or TurboID for BioID (step 1), which biotinylate nearby proteins upon incubation of engineered cells with biotin in culture (step 2). Control lines can express the labelling enzyme fused to a non-specifically localized control bait such as green fluorescent protein or a localization signal specific for a non-target organelle. In the case of APEX, the addition of H2O2 generates short-lived biotin-phenol free radicals that react with nearby biomolecules. Following labelling, a streptavidin pull-down step enriches for labelled proteins, which can then be identified by mass spectrometry (MS) (step 3). High-confidence proximity interactors are determined by comparing preys with proteins isolated to control lines using such tools as SAINT (Significance Analysis of INTeractome) (step 4). b | Organellar components can be elucidated using bait-centric or prey-centric analyses. Ratiometric quantification of baits, for example using isotopic labelling approaches with baits in and outside the organelle, or hierarchical clustering of multiple baits can be used to identify enrichment of proteins within an organelle of interest in a bait-centric manner. Alternatively, extensive proximity interaction networks can be elucidated using multiple baits and prey-centric analyses including Pearson’s correlation and factorization approaches such as non-negative matrix factorization (NMF). LC-MS/MS, liquid chromatography with tandem mass spectrometry; m/z, mass to charge ratio; SILAC, stable isotope labelling by amino acids in cell culture; TMT, tandem mass tagging; t-SNE, t-distributed stochastic embedding; LFQ, label-free quantitation.
