Fig. 4 |. Generic fluorescence immunocytochemistry proteomics workflow.
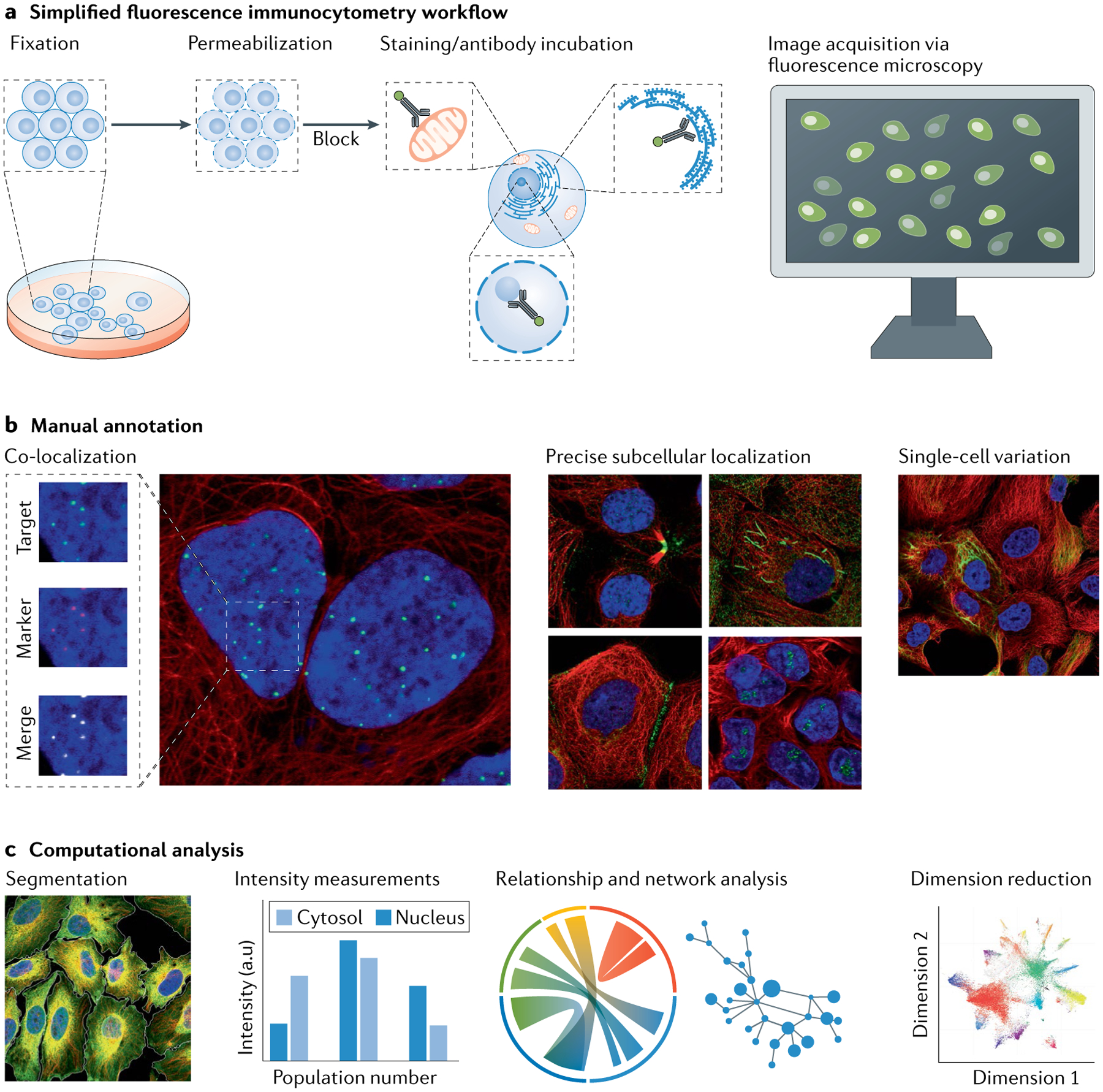
a | Cells are fixed using either cross-linking agents or organic solvents, such as aldehydes or alcohols. Cross-linkers generally outperform organic solvents for preserving subcellular structures but may reduce antigen retrieval for certain proteins. Permeabilization of cells with detergents such as digitonin or Triton X-100 extracts lipids from the cell membrane and allows for penetration of affinity agents such as antibodies into the cells. Blocking buffer containing serum or albumin helps reduce unspecific labelling before addition of affinity reagents. Cells are then counterstained with different organelle probes before imaging. Image acquisition is typically performed using confocal microscopy and an optional embedding step can be used for archiving of the samples. b,c | After image acquisition, the image data are annotated to assign protein location to subcellular structures. b | Manual inspection of images can be used to acquire qualitative data. For obvious staining patterns easily interpreted by eye, qualitative annotation might be sufficient if the data sets are small. c | For large image data sets, computational analysis is required. Computational strategies enable high-throughput collection of quantitative, morphological and comparative information, such as algorithms for segmentation and intensity measurements. Such quantitative data can then be used to generate networks and dimension reduction plots. Multi-label patterns and fine structures stained in a subset of cells may require manual annotation or more advanced computational analysis. Relationship and network analysis in part c adapted from REF.11, AAAS. Dimension reduction plot in part c adapted from REF.180, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
