Figure 2.
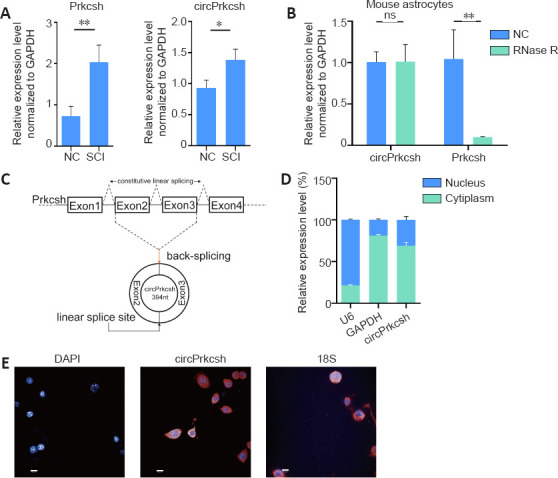
Circular RNA verification of circPrkcsh.
(A) The levels of Prkcsh and circPrkcsh expression were significantly different between tissue from the SCI and sham groups at 3 days post-surgery, based on RT-qPCR analysis. (B) The mRNA levels of circPrkcsh and Prkcsh in astrocytes with or without RNase R treatment were detected using RT-qPCR, and were then normalized to Gapdh. (C) Schematic illustration showing that circPrkcsh is formed by Prkcsh exon 2–3 circularization (black arrows). RT-qPCR was performed to confirm the presence of circPrkcsh, followed by Sanger sequencing. Head-to-tail circPrkcsh splicing sites are marked with a red arrow. (D) The circPrkcsh expression in different cell fractions was detected using RT-qPCR. U6 was used as the nuclear RNA internal control, and GAPDH was used as the cytoplasmic RNA control. Data are expressed as the mean ± SD based on triplicate independent experiments. *P < 0.05, **P < 0.01 (Student’s t-test). (E) RNA FISH revealed that circPrkcsh was predominantly located in the cytoplasm. DAPI was used to label the nuclei, while Cy-3 was used to label the circPrkcsh probes. The cellular localization of circPrkcsh was indicated using FISH with junction-specific probes. Scale bars: 50 µm. DAPI: 4′,6-Diamidino-2-phenylindole; FISH: fluorescence in situ hybridization; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; NC: normal control; ns: not significant; Prkcsh: protein kinase C substrate 80K-H; RT-qPCR: reverse transcription-quantitative polymerase chain reaction; SCI: spinal cord injury.
