Figure 1. Principles of combinatorial and temporal coding for stimulus-specific gene expression and the layers of the stimulus-responsive network enabling it.
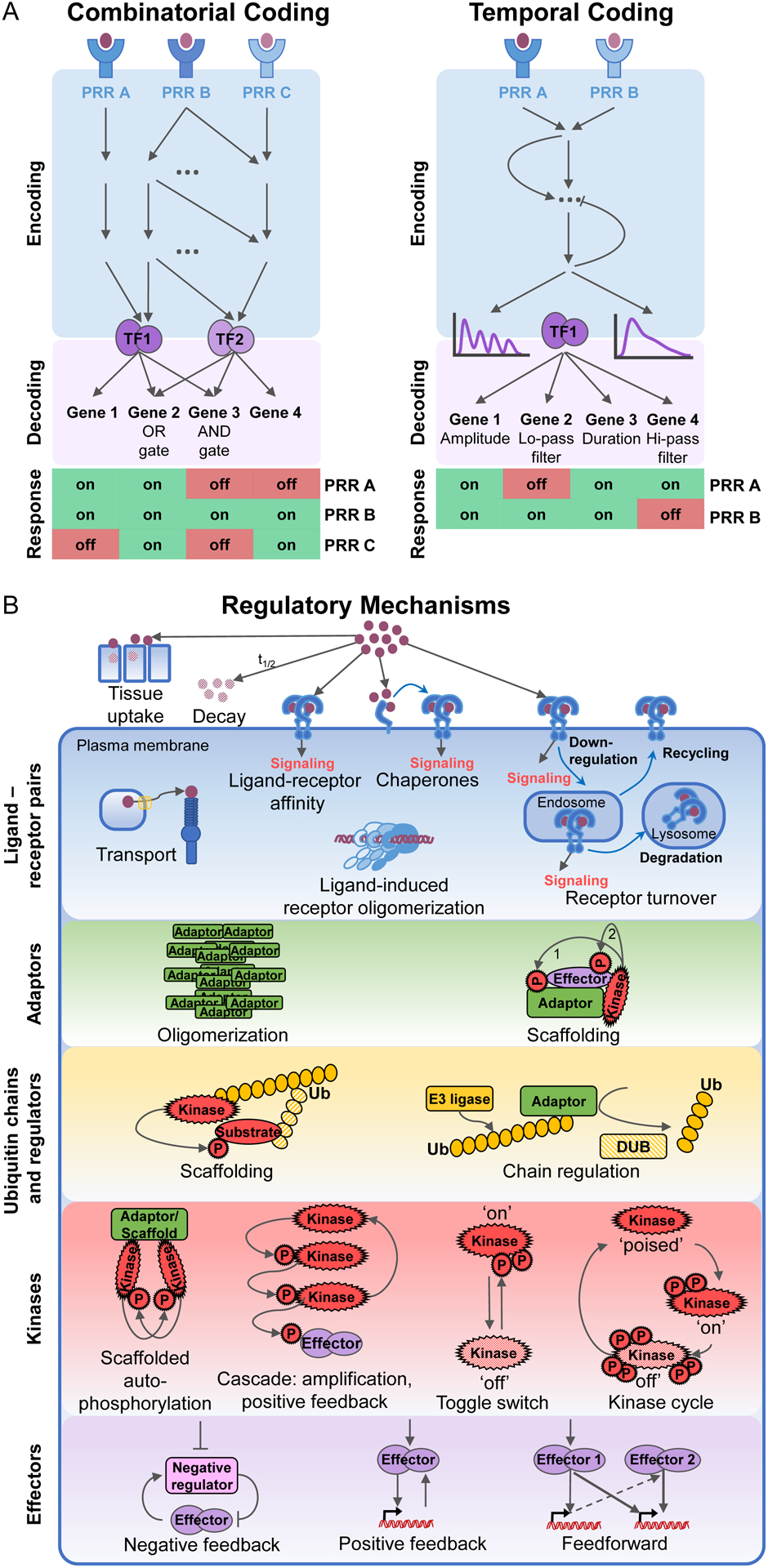
(A) In combinatorial encoding, receptor proximal mechanisms activate stimulus-specific effector combinations. Response genes decode this through Boolean logic gate-based regulation. Stimulus-specific activity profiles of single effector modules allow temporal encoding of information, which is decoded by gene-specific mechanisms. PRR: pattern recognition receptor; Lo-pass filter: low pass filter; Hi-pass filter: high pass filter.
(B) Regulatory strategies controlling immune sentinel cell responses in the signaling network. The stimulus-responsive network consists of multiple functionally distinguishable signaling layers: (1) ligand-receptor pairs, (2) receptor-associated adaptors, (3) ubiquitin signaling layer, (4) kinase hubs, (5) SDTF modules and other effectors. Biochemical and cell biological mechanisms are shared between the diverse components of each signaling layer that together enable stimulus-specific coding. Ligand-receptor interactions in different subcellular locations are regulated through ligand stability, diffusion, transport, binding chaperones, and receptor transport and turnover. Adaptor proteins scaffold the assembly of larger signaling complexes. The kinetics of adaptor complex oligomerization are a key determinant of downstream signaling. Ubiquitin chains of different linkages pass on, fine-tune, and eventually downregulate signaling. They are regulated by the complex interplay of E3 ligases and DUBs. Signaling converges on three key kinase families, which are activated through scaffolded autophosphorylation or kinase cascades with toggle switch or kinase cycle mechanisms. The effector molecules activated by kinases are transcription factors inducing gene expression or posttranscriptional regulators. They are regulated through negative and positive feedback and feedforward loops. DUBs: deubiquitinases, Ub:ubiquitin, P: phosphoryl group.
