Abstract
Toll-like receptors (TLRs) are the important mediators of inflammatory pathways in the gut which play a major role in mediating the immune responses towards a wide variety of pathogen-derived ligands and link adaptive immunity with the innate immunity. Numerous studies in different populations across the continents have reported on the significant roles of TLR gene polymorphisms in modulating the risk of colorectal cancer (CRC). CRC is one of the major malignancies affecting the worldwide population and is currently ranking the third most common cancer in the world. In this review, we have attempted to discuss the structure, functions, and signaling of TLRs in comprehensive detail together with the role played by various TLR gene SNPs in CRC susceptibility.
1. Introduction
Colorectal cancer (CRC) is one of the commonly diagnosed cancers representing about 10% of all cancers worldwide and is currently ranked as the third most common cancer in men and second most in women worldwide [1–3]. CRC usually arises from the inner wall of the intestines and does involve the colon rectum and appendix [4].
CRC is among few solid tumors in which inflammation acts as a founder effect and plays a key role in the carcinogenesis mechanism in transforming the normal cells into the malignant ones [5, 6]. In addition, numerous researchers have demonstrated a positive association between various innate immune system mediators and bacterial toxins with the development of CRC have also been found [7–9]).
CRC has a multifarious etiology and is usually classified as sporadic, inherited, or familial in their origin and occurrence [10]. With respect to the risk factors, age, sex, and race, the inflammatory diseases are recognized as the prime ones for the development of the CRC [11]. CRC initiation and progression follow a multigene and multistage genetic instability model called the Vogelstein model of carcinogenesis [12]. Numerous gene mutations (suppressing and activating), gene silencing, and genetic polymorphisms have been shown to play their role in driving the carcinogenesis pathway, with each stage having a unique molecular fingerprint of genetic variations [10, 11].
Currently, three pathways for the genetic instability have been identified for the CRC initiation, development, and progression which are as follows: chromosomal instability (CIN) pathway, microsatellite instability (MSI) pathway, and CpG island methylator phenotype (CIMP) pathway [11, 13, 14].
Toll-like receptors (TLRs) have been identified as the important mediators of inflammatory pathways in the gut which serve as crucial regulators in maintaining the balance between commensal bacteria in the gut and the mucosal immune system [15–20]. Numerous studies across the populations and across the continents have reported on the significant roles of TLR gene polymorphisms modulating the risk of CRC [21–28].
In this review, we aim to sketch an in-depth picture about the importance of TLR functions, their signaling, and the role genetic polymorphisms of some of the important and common TLR genes do play in the colorectal cancer susceptibility.
2. Toll-Like Receptors (TLRs)
Toll-like receptors (TLRs) are evolutionarily conserved receptors belonging to the family of pattern recognition receptors (PRRs) which play a vital role in immune responses especially pathogen recognition by the extracellular matrix [18, 29–31]. TLRs are directly involved in the regulation of inflammatory reactions and activation of the innate or adaptive immune responses for the elimination of infectious pathogens and cancer debris [29, 32–34].
TLRs hold a key position in the first line of defense against pathogens because of their ability to recognize the conserved pathogen-associated molecular patterns (PAMPs), conserved structures of the pathogens, or the damage caused by the pathogens within the host [35–38].
Pattern recognition receptors (PRRs) make up a key component of the innate immunity because of their ability to initially sense the exposure to infection and elicit an intracellular signaling cascade for the eventual elimination of the pathogen and infected cell thereof [18]. All PRRs are germline-encoded proteins which can recognize wide varieties of alien molecules (e.g., lipid, carbohydrate, peptide, and nucleic acid) commonly found in pathogens but distinct from the host molecules and thus are commonly referred to as PAMPs [39]. Activation of PRRs results in the downstream transcriptional activation and expression of numerous inflammatory mediators [18, 35, 36, 38]. In addition, PRR signaling also leads to the triggering of various processes involved in autophagy, cell death, cytokine processing, and phagocytosis [40, 41].
Currently, based on their composition and structure, PRRs are categorized into five classes depending on their primary functions, position, specificity for ligands, and evolutionary associations as follows:
Toll-like receptors (TLRs)
AIM2-like receptors (ALRs)
C-type lectin receptors (CLRs)
Retinoic acid-inducible gene- (RIG-) I-like receptors (RLRs)
TLRs and CLRs function as membrane-bound receptors and exist as transmembrane proteins, while others occur as cytosolic proteins [18, 35, 42]).
PRRs have been found to be expressed not only in specialized immune response cells like macrophages, neutrophils, dendritic cells (DKs), natural killer (NK) cells, mast cells, basophils, and eosinophils but also on other cells as well [32, 35].
Among the numerous molecular patterns which PRRs can recognize as foreign and be able to elicit an innate immune response, four are characterized till date which are as follows: pathogen-associated molecular patterns (PAMPs), damage/danger-associated molecular patterns (DAMPs), microbe-associated molecular patterns (MAMPs), and xenobiotic-associated molecular patterns (XAMPs). Most of the PRRs after getting activated by the molecular patterns from the pathogens cause the upregulation of the inflammatory response gene especially proinflammatory cytokines and type I interferons [18, 35, 42]. While TLRs and CLRs are sensitive to the molecular patterns located outside the cell because of their surface expression, others respond to the pathogen-derived patterns intracellularly.
2.1. TLRs: History, Structure, and Functions
The discovery of the TLR receptors dates to the identification and functional characterization of the toll gene and its protein in Drosophila [43]. TLRs received their name because of their similarity to the toll protein in serving as the cell surface receptors [44], which play a role in providing the immunity against the fungal and gram-positive bacterial infections [45] together with their role in embryonic development and dorsoventral polarization [35, 46–49]).
TLRs are categorized as a type of type I integral transmembrane proteins, usually consisting of three domains—an N-terminal domain (NTD) located outside the membrane, a middle single helix transmembrane domain traversing the membrane, and a C-terminal domain (CTD) located towards the cytoplasm [50]. N-terminal domain makes up an ectodomain to serve as the ligand recognition site to the variety of PAMP, while CTD is involved in the interactions with various signal transduction adaptors for the initiation of downstream signaling through its toll-IL-1 receptor (TIR) homologous domain [18, 35, 42, 48, 50]. The ectodomain has a folded solenoid structure (resembling a horseshoe) of about 19-25 highly conserved short tandem leucine-rich repeat (LRR) motifs, which provide the much-needed specificity and recognizability to the TLRs for PAMPs/DAMPs [36, 51]. Each LRR repeat sequence of the TLRs contains 24-29 amino acids in a sequence pattern of xLxxLxLxx [35, 52]. In addition, the NTD does contain glycan moieties which serve as the actual binding sites for various pathogen-derived ligands [29, 53]. Figure 1 provides a comparison between the two receptors.
Figure 1.
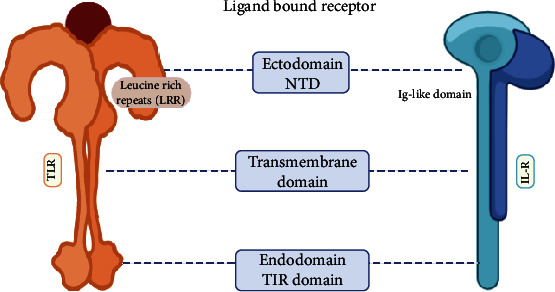
A comparative structure of TLR and IL-R.
2.2. TLR Family Members
TLRs are further subdivided into two groups, depending upon the number of cysteine clusters present within the extracellular LRR motif as follows: vertebrate type (V-Type) and protostome type (P-Type). V-Type TLRs possess a single cluster of cysteine or CF motif on their CTDs (LRRCT) and hence also referred to as Single Cysteine Cluster TLRs (sccTLRs), and P-Type TLRs have multiple (two or more) cysteine clusters or CF motifs either on their LRRCT or on the N-terminal (LRRNT) domain [54, 55].
The TLR family plays a critical role in the immune system of an individual, be it an insect or a human being. They are one of the best-characterized PRR families, which function to recognize the self and nonself antigens, detect various pathogens, bridge the innate and adaptive immunity, regulate cytokine production, and regulate proliferation and survival of the host cell [18, 35, 48, 56].
To date, 222 TLRs have been identified in invertebrates and 28 TLRs in vertebrates, with the highest number of TLRs in an individual found in the vertebrate fish of teleost which possesses 21 TLR molecules—TLR1-5, TLR7-9, TLR13-14, TLR18-23, and TLR25-28 [35, 42, 48, 55, 57, 58] (Table 1). In mammals, a total of 13 TLRs (TLR1-13) have been found—however, humans possess only 10 (TLR1-10) of them while mouse possesses all with TLR10 being nonfunctional [35, 42, 48, 57]. Much of the ligands and the signaling pathways of the TLR1-9 and 11 are known, while the biological role of TLR10, 12, and 13 still is not clear. In humans, TLR4 was the first of the TLR family to be discovered and reported to regulate the inflammatory responses [59], while TLR3 is the most ancient one which belongs to the viral TLRs like TLR7-9 [35, 53].
Table 1.
Characteristics of different human TLRs.
| TLR member | TLR expression | TLR coreceptor | Gene & its location | Origin | TLR active form | Signaling adaptors |
|---|---|---|---|---|---|---|
| TLR1 | Cell membrane | TLR2 |
TLR1-6-10
4p14 |
Nonviral | TLR1-TLR2 | BCAP, TIRAP, MyD88, SCIMP |
| TLR2 | Cell membrane | TLR1, 2, 6 & 10 CD14, CD36, integrin, RP105, MBL, LBP |
TLR2
4q31.3 |
Nonviral | TLR1-TLR2 TLR2-TLR2 TLR2-TLR6 TLR2-TLR10 |
BCAP, TIRAP, MyD88, SCIMP |
| TLR3 | Endoplasmic reticulum, lysosomal membrane | CD14, Mex3B |
TLR3
4q35.1 |
Viral | TLR3-TLR3 | SARM, SCIMP, TRIF, TICAM1 |
| TLR4 | Cell membrane, endoplasmic reticulum, lysosomal membrane | MD2, LY96, CD14, CD36, LBP, RP105 |
TLR4
9q33.1 |
Nonviral | TLR4/MD2-TLR4/MD2 TLR4-TLR6 |
BCAP, TIRAP, MyD88, SARM, SCIMP, TICAM1, TICAM2 |
| TLR5 | Cell membrane |
TLR5
1q41 |
Nonviral | TLR5-TLR5 | MyD88, TICAM1 | |
| TLR6 | Cell membrane | TLR2, CD36, LBP |
TLR1-6-10
4p14 |
Nonviral | TLR2-TLR6 TLR4-TLR6 |
BCAP, TIRAP, MyD88, SCIMP |
| TLR7 | Cell membrane, endoplasmic reticulum, lysosomal membrane | CD14 |
TLR7
Xp22.2 |
Viral | TLR7-TLR7 | MyD88 |
| TLR8 | Endoplasmic reticulum, lysosomal membrane |
TLR8
Xp22.2 |
Viral | TLR8-TLR8 | MyD88 | |
| TLR9 | Endoplasmic reticulum, lysosomal membrane | CD14 |
TLR9
3p21.2 |
Viral | TLR9-TLR9 | TIRAP, MyD88, SCIMP |
| TLR10 | Cell membrane, endoplasmic reticulum, lysosomal membrane |
TLR1-6-10
4p14 |
Nonviral | TLR1-TLR10 TLR2-TLR10 TLR10-TLR10 |
MyD88 |
Depending upon their functionality and location in the host cell, TLRs are further categorized into two types [18, 35, 38, 48, 56] as follows:
Cell membrane TLRs, which are expressed in their active form on the cellular surface, with NDT ectodomain facing outside for sensing and binding with the alien ligands. They include TLR1, 2, 4, 5, 6, and 10. In their active signal-transducing form, they exist as dimers—TLR1-TLR2, TLR2-TLR6, and TLR2-TLR4/5/10 [60]
Intracellular TLRs, which are expressed within the host cells on the organelle biomembranes like endoplasmic reticulum (ER), endosomes, and lysosomes. They include TLR3, 7, 8, and 9 [52, 61]
3. TLR Ligands
TLRs are predominantly expressed in the host antigen-processing cells (APCs) which play crucial roles in recognizing the pathogenic infection and deciding their fate [62, 63]. TLRs have evolved to recognize a diverse array of PAMPs from a range of microorganisms including bacteria, protozoa, fungi, and viruses [20, 52, 53, 62, 63]. For almost each and every kind of major biomolecules derived exogenously from the pathogens (PAMPs) be it a protein, carbohydrate, lipid, or a nucleic acid, there exist one or more TLRs in the host cell for its binding and recognition as a foreign entity (Figure 2) [20, 52, 63]. Similarly, TLRs also do bind with various endogenous ligands (host-derived DAMPs) like plasma membrane constituents, heat shock proteins (HSPs), and organelle nucleic acids like mitochondrial DNA (mt DNA) [64].
Figure 2.
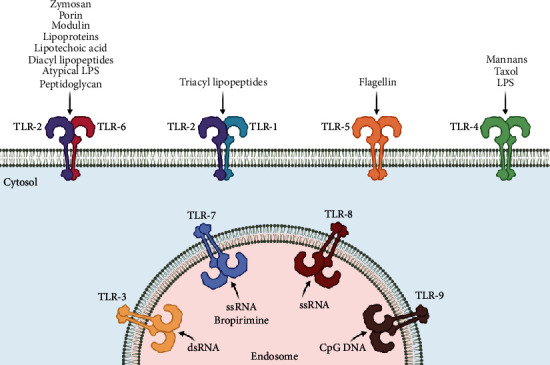
Detection of PAMPs by various TLRs.
However, TLR2 and TLR4 are the most efficient of the PRRs in being able to recognize and consequently get activated by a wide variety of ligands [20, 63] (Table 2). Some of the TLRs are highly specific in recognizing and binding to some particular ligands, like TLR2 for lipoproteins, lipotechoic acid (LTA), peptidoglycans (PG), zymosan and porin [65, 66], TLR3 for dsRNA derived from viruses [67], TLR4 for gram-negative bacteria-derived lipopolysaccharides (LPS) and plant-derived taxol [64, 68], TLR5 for the gram-negative bacterial flagellin protein [69], and TLR7 for antiviral synthetic compounds—imidazoquinolines and loxoribine [70, 71].
Table 2.
TLR recognition of microbial components.
| Origin | Exogenous | Endogenous | ||||
|---|---|---|---|---|---|---|
| Pathogen | Bacteria | Fungus | Parasites | Viruses | Synthetic compounds | Host |
| Derivatives | LPS Atypical LPS Diacyl lipopeptides Triacyl lipopeptides Lipoteichoic acid Peptidoglycan Glycolipids Porins Lipoarabinomannan Flagellin CpG-DNA |
Zymosan Taxol Phospholipomannan Mannan Glucuronoxylomannan |
tGPI-mutin Glycoinositolphospholipids Hemozoin Profilin-like molecule |
DNA dsRNA ssRNA Envelope proteins Hemagglutinin protein |
Bropirimine Guanosine analogs Imidazoquinolines Loxoribine |
Heat-shock protein 60, 70 Fibrinogen Hyaluronan Hemagglutinin Chromatin-IgG complexes mtDNA |
|
| ||||||
| TLR recognition | TLR2, 4, 5, 6, 9 | TLR2, 4, 6 | TLR2, 4, 9, 11 | TLR2, 3, 4, 9 | TLR7 | TLR 4 |
4. TLR Signaling Pathways
On activation by binding of the ligand to the LRR domain of TLR, the signal needs to be transferred to the internal environment of the cell. For TLRs, various adaptor molecules have been identified which get recruited to the CTD domain of the activated and/or dimerized receptor to pass the signal downstream of the cell [31, 72, 73]. The important adaptors which are critical for the functioning of the TLR signaling are identified by the possession of the toll-IL-1 receptor (TIR) domains in their structure and are categorized as follows:
Myeloid differentiation primary-response protein 88 (MyDD88)
TIR domain-containing adaptor protein (TIRAP or Mal)
TIR domain-containing adaptor inducing IFN-β (TRIF, TICAM1)
TRIF-related adaptor molecule (TRAM, TICAM2)
B-cell adaptor for phosphoinositide (BCAP)
Sterile α- and armadillo-motif-containing protein (SARM)
Since TLRs belong to the class I transmembrane receptor family, the binding of the ligand induces a dimerization which in turn activates them [20, 31, 35, 52, 63]. The actual signaling cascade is dependent upon the type of ligand, interacting TLR, and the downstream adaptor molecule engaged for the signaling pathway. However, TLR signaling is generally divided into two distinct pathways based on the recruitment of either MyD88 or TRIF for the downstream signaling cascades [18, 31, 35]. Figure 3 sketches the essential dynamics of the two pathways of TLR signaling.
Figure 3.
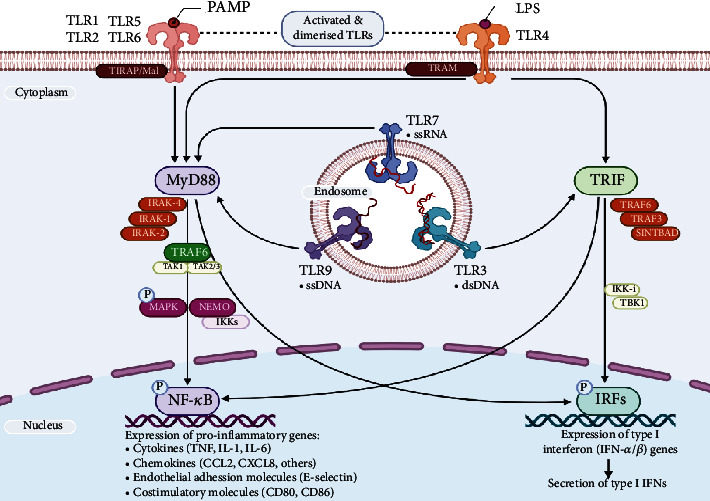
Dynamics of the toll-like receptor signaling.
MyD88-dependent pathway
It is utilized by all TLRs (except TLR3) which eventually lead to the enhanced expression of proinflammatory cytokine genes. MyD88 recruitment to the activated CTD domain of the TLRs is facilitated by two important adaptor molecules—Mal and TIRAP. Mal adaptor protein is utilized by all kinds of TLR receptors to recruit MyD88, while TLR1, 2, 4, and 6 require additional association with TIRAP molecule to facilitate the contact with MyD88 [52].
TRIF-dependent pathway
It is utilized by TLR3 and TLR4 to trigger the enhanced expression of interferon type-1 genes. TLR3 utilizes it in response to the binding of viral dsRNA by recruiting TRIF directly, while TLR4 requires another adaptor protein, TRAM, for the activation of TRIF [18].
4.1. Effect on Carcinogenesis
The specific ligand activation of the TLR signaling has dual and contrasting functions on the carcinogenesis pathways. On the one hand, TLR signaling has been shown to promote the carcinogenesis through proinflammatory, antiapoptotic, proliferative, and profibrogenic signals within the tumor microenvironment or tumor cells themselves; while on the other hand, TLR signaling induces a highly sensitive and effective tumor immunosurveillance by activating specific immune cells like dendritic cells (DCs), natural killer cells (NKs), and cytotoxic T lymphocytes (CTLs), all which have antitumorigenic activity [74]. Figure 4 summarizes the two faces of the TLR signaling in carcinogenesis.
Figure 4.
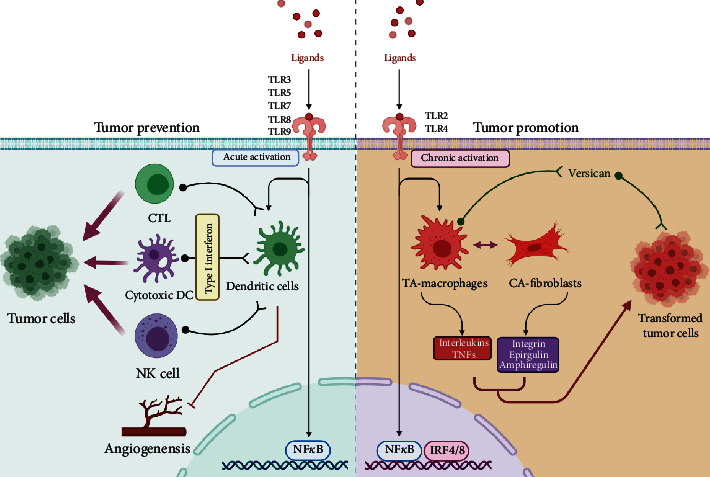
The two faces of the TLR signaling in carcinogenesis.
Activation of one class of TLRs in particular TLR3, 5, 7, 8, and 9 does play a role in providing the antitumor immunity by modulating the function of immune tolerance via type I interferon [18, 31]. Primary modulation occurs via DC activation, which otherwise are chief mediators of immunosurveillance [75]. Within the tumor environment, DCs serve as the chief antigen-presenting cells (APCs) by expressing large number of PRRs including TLRs. DCs mediate the antigen processing and presentation for both innate and adaptive immune responses. On activation, DCs exert their effective modulating role on the key effectors like NKs and CTLs in a type I interferon-dependent manner [74, 76]. Additionally, TLR-induced expression of alpha IFNs is known to limit tumorigenesis by suppressing the angiogenesis [77, 78].
On the other hand, ligand-specific low and chronic stimulation of TLR2 and 4 leads to the tumor-promoting effects by enhancing inflammatory and antiapoptotic signals. The central role in both these effects is played by the key transcription factor NF-κB [74]. NF-κB has two-prong effects on the cellular environment—on the one hand, the NF-κB-dependent TLR pathway enhances the overexpression of numerous proinflammatory molecules including interleukin- (IL-) 1b, tumor necrosis factor a (TNFa), and IL-6 [18, 31, 79–81]; while on the other hand, it leads to the increased expression of many antiapoptotic genes in addition to the restriction of proapoptotic pathways mediated by c-Jun kinase [82, 83]. Moreover, within the tumor microenvironment, two important TLR expressing cells, i.e., tumor-associated macrophages (TAMs) and cancer-associated fibroblasts (CAFs), have been reported to provide survival signals for tumor growth [74]. Also, within the tumor microenvironment, there is an increased expression and secretion of versican, a large ECM proteoglycan, which is an important component of tissue inflammation as a result of infection or injury [84]. Versican results in further enhanced activation of TLR signaling thereby amplifying and fueling the tumor-promoting inflammatory signals furthermore [74].
5. Toll-Like Receptor Polymorphism and Colorectal Cancer
TLRs are important cogwheels of the immune machinery. Because of their ability to recognize and bind with a wide variety of PAMPs (bacteria, parasites, fungi, and viruses) and DAMPs, they act as important regulators of the immune responses and bridge the innate with adaptive immune responses in the host [20, 31, 35, 52, 63, 85]. Majority of the immune response elicited by the TLRs is due to the highly specific ectodomain containing a germ line encoded and highly conserved LRR region [18, 31, 53]. Much of the ligand specificity of each TLR is because of this LRR region [18].
Various TLRs play a critical role in innate immunity by responding to the various PAMPs (from the pathogenic bacteria, viruses, or fungi) which drive the cellular response producing proinflammatory cytokines, chemokines, and other mediators, thus playing an important role in inflammatory reactions and activation of the adaptive immune pathways as well [86–88]. TLR-induced proinflammatory responses are considered as the first line of host defense, not only working against the pathogens but also accelerating the healing process to restore the immune homeostasis [89].
It has been demonstrated that any dysregulation in the TLR signaling, be it in ligand recognition and binding or in downstream signaling, has a founder effect in various immunopathological diseases like autoimmune diseases, allergy, arthritis, atherosclerosis, tuberculosis, rheumatic disorders, pancreatitis, and malignancies [22, 24, 88–93].
Any genetic variations (GVs) be it a mutation or a polymorphism which affects the TLR genes thereby affects the structure of TLRs especially their ligand-binding domains (NTD-LRR) and intracellular (CTD). TIR domain may play a role in modulating the incidence, the severity, and the outcome of many immune-related diseases [89, 94]. With the advent of technology and easy sequencing techniques, several studies have demonstrated the existence of numerous genetic polymorphisms affecting almost all TLR genes which have influence on the immune responses of the host towards the pathogenic infection or the consequent PAMPs and DAMPs [19, 22, 85, 89, 94].
In colorectal carcinogenesis, inflammation has been demonstrated as a major risk factor, with the involvement of different driver molecules like NF-κB, interleukins, tumor suppressor proteins, oncogenes, cyclooxygenase, nitric oxide synthases, and growth factors including TLRs [85, 86, 95]. Numerous studies have linked the dysregulation of the TLR expression to the colorectal carcinogenesis [16, 26, 27, 96–100], and now, the evidence is mounting for the role of genetic polymorphisms in modulating the risk of CRC in various populations as well [17, 19, 21, 22, 24, 28, 49, 89, 94].
Of all the TLRs that are functional in humans, the single nucleotide polymorphisms (SNPs) of five TLR genes—TLR2, 3, 4, 5, and 9—have been reported to be involved in modulating the risk of CRC extensively [17, 21, 22, 25, 28, 49, 101, 102]. SNPs in any gene results in the change of amino acid sequence (missense/frameshift) which alters the structure and activity of the involved protein [103]. Numerous SNPs in the TLR gene especially in the NTD-LRR domain result in the disruption of the pathogen recognition ability as well as immune functions of these receptors (see Table 3) [89, 94].
Table 3.
Summary of various TLR SNPs reported in colorectal cancer susceptibility.
| Gene | Location | SNP | Major/minor allele |
|---|---|---|---|
| TLR2 | 4q32 | Delta22 | Ins/del |
| GT repeats | GT | ||
| rs1898830 | A/G | ||
| rs4696483 | C/T | ||
| rs3804099 | T/C | ||
| rs7656411 | T/G | ||
| rs5743704 | C/A | ||
| rs5743708 | G/A | ||
|
| |||
| TLR3 | 4q35 | rs5743305 | T/A |
| rs11721827 | A/C | ||
| rs3775290 | A/G | ||
| rs3775291 | G/A | ||
| rs3775292 | C/G | ||
|
| |||
| TLR4 | 9q32-q33 | rs10759932 | T/C |
| rs1927911 | C/T | ||
| rs5030728 | G/A | ||
| rs11536898 | C/A | ||
| rs12377632 | T/C | ||
| rs11536889 | G/C | ||
| rs1554973 | T/C | ||
| rs4986790 | A/G or A/T | ||
| rs4986791 | C/T | ||
|
| |||
| TLR5 | 1q41 | rs5743836 | A/G |
| Rs5744168 | G/A | ||
| rs5744174 | A/G | ||
| rs2072493 | T/A or T/C | ||
|
| |||
| TLR9 | 3p21.2 | rs187084 | A/G or A/T |
| rs352140 | C/A or C/G or C/T | ||
5.1. TLR2
The TLR2 gene is located on the long arm of chromosome 4 (4q31.3), coding for a protein of 784 amino acid residues of 89.9 kDa molecular weight [104].
Among the various SNPs that have been reported in TLR2, one is the most studied and reported in CRC susceptibility—TLR2 Arg753Gln (+2258G/A, rs5743708) SNP [22, 49]. TLR2 rs5743708 SNP occurs in the TIR domain of the TLR2 and has been found to result in the impaired NF-κB signaling and decreased secretion of cytokines in response to the PAMPs [89].
Recently, another TLR2 genetic variation, a 22 bp deletion at position -196 to -174 of the promoter region, has been also reported in CRC (Delta22). This GV has been reported to cause the reduction in the transcription resulting in the reduced expression of the receptors in the cell [5, 105]. Zhu et al. [105] had reported that a significant association was found between the TLR2 −196 to −174 del and cancer risk; however, it was restricted to only a subgroup of Caucasian and south Asians and not among east Asians, while Proenca et al. Proenca et al. [5] reported that TLR2 -196 to -174 del polymorphism results in the increase in the TLR2 mRNA expression which is associated with a higher risk of developing CRC. In contrast, Pimentel-Nunes et al. [27] reported that TLR2 (+597T/C, rs3804099) resulted in a fivefold decreased risk of CRC. Furthermore, Boraska et al. [106] found the unique microsatellite GT repeat polymorphism in the intronic region of the TLR2 gene to be associated with the CRC in the Croatian population. This repeat genetic variation is in the intron 2, 100 bp upstream of the translation start site, where GT repeats vary from 12 to 28 [107]. Based on the repeat structure, three different alleles have been categorized: (GT)16 or less as S, (GT)17-22 as M, and (GT)23 or more as L allele, and are present in different populations in different frequencies [107].
5.2. TLR3
The TLR3 gene is located on the long arm of chromosome 4 (4q35.1), coding for a protein of 904 amino acid residues of 103.8 kDa molecular weight [108].
About 10 SNPs have been reported to span an entire coding region of the TLR3 gene, and among these five SNPs, rs5743305, rs11721827, rs3775290, rs3775291, and rs3775292 have been extensively studied and reported to modulate the risk of cancers including CRC [17, 109]. Wang et al. [109] in their meta-analysis reported the association of only two TLR3 polymorphisms, rs11721827 and rs3775292, with the risk of CRC. Additionally, TLR3 rs3775291 was reported to be associated with CRC-specific survival, with patients having TT genotype having a 93% increased risk of death in comparison to CC ones [110].
5.3. TLR4
The TLR4 gene is located on the long arm of chromosome 9 (9q33.1), coding for a protein of 839 amino acid residues of 95.6 kDa molecular weight [111].
Of all the present SNPs in the TLR4 gene, two nonsynonymous SNPs (+896A/G, rs4986790 and +1196C/T, rs4986791) located within the exon 3 of a gene are known to induce substitution of amino acids Asp299Gly and Thr399Ile, respectively [97, 112, 113]. The substitution of Asp299Gly disrupts the normal structure of the extracellular region of the TLR4, which may cause decreased ligand recognition or protein interaction and decreased responsiveness to lipopolysaccharide (LPS) [5, 114]. This change affects the cell surface expression of the TLR4 protein and is reported to cause inflated inflammatory response leading to severe tissue destruction [105].
TLR4 Asp299Gly and Thr399Ile are known to cosegregate in 10% of the Caucasian populations and are reported to have positive correlation with susceptibility to several diseases [22, 49]; but neither SNP is present in Asian populations [21, 112]. Numerous studies and meta-analyses have linked TLR4 SNPs with the increased risk of CRC [22, 86, 113, 115]. In a recent meta-analysis, Sheng et al. [113] demonstrated that GG genotype of TLR4 rs4986790 and TT genotype of TLR4 rs4986791 SNPs are correlated with the risk of CRC. A previous meta-analysis by Zhu et al. [105] had established a significant association of TLR4 SNPs with cancer especially in Caucasian and south Asian populations. In addition, a recent study by Moaaz et al. [115] reported that both these SNPs are associated with the risk of CRC in the Egyptian population. In addition, a recent study in the Danish population did implicate the TLR4 and TLR2 and their interaction with the meat-rich diet with the CRC risk [116].
5.4. TLR5
The TLR5 gene is located on the long arm of chromosome 1 (1q41), coding for a protein of 858 amino acid residues of 97.8 kDa molecular weight [117]. Of the many studied genetic polymorphism in the TLR5 gene, two SNPs of the TLR5 gene are the most important ones which have been linked with gastrointestinal cancers including CRC, which are TLR5 rs2072493 and rs5744174 [19, 22, 89].
The rs5744174 SNP leads to the substitution of leucine to phenylalanine at 616 amino acid residues of TLR5, thereby affecting the ligand recognition capacity of the receptor, contributing to the increased inflammation and hence increase the risk of cancers [19, 28]. Since TLR5 holds a key to identify among all TLRs in being the unique receptor for bacterial flagellin protein recognition, the change in ligand binding does affect the downstream signaling and eventual secretion of IL-1β and IL-6. Both are key inflammatory molecules playing a direct role in inflammatory pathways of CRC carcinogenesis [28, 89]. Recently, Klimosch et al. [118] reported that these two TLR5 SNPs did disparately affect the risk of CRC with rs2072493 being associated with worse survival while rs5744174 with better survival.
5.5. TLR9
The TLR9 gene is located on the short arm of chromosome 3 (3p21.2), coding for a protein of 1032 amino acid residues of 115.8 kDa molecular weight [119]. In the TLR4 gene, two polymorphisms have been identified to play a role in the CRC tumorigenesis risk [19, 22, 89]. TLR9 is a unique receptor for the unmethylated DNA derivatives from the viruses and bacteria [24, 92]. Although the increased expression of TLR9 has been reported in the advanced CRC [120, 121], its two SNPs—TLR9 rs187084 and rs352140—have lately been found to be associated with the inflammatory diseases which are regarded as the risk factor for CRC as well [19, 28].
6. Toll-Like Receptors and Therapeutics
CRC is one of the malignancies in which inflammation is considered as the pivotal risk factor [10, 23], coupled with the key role TLRs play in the inflammation-driven carcinogenesis [5, 6, 15, 16, 29, 53, 95]; targeting various TLRs with the various agonists, antagonist, and vaccine adjuvants is being currently explored as one of the strategies of cancer therapeutics [23, 122, 123].
Recently, it has been established that TLR agonists do exhibit varied therapeutic benefits as antitumor agents via their ability to activate immune cells within the tumor microenvironment on the one hand and increase the expression of cytokines on the other. The dual effects help in the activation of antitumorigenic dendritic cells, cytotoxic T lymphocytes, and natural killer cells which play an active role in the suppression of oncogenic signaling pathways ([51, 74, 75, 122]; ). Usually, there are two chief strategies which are utilized in cancer therapeutics in achieving the TLR inhibition as follows: (a) preventing the ligand binding to the TLR receptor and (b) impeding the function of various proteins located downstream of the TLR signaling [23, 51, 122, 124, 125].
The role of TLR signaling in the colorectal carcinogenesis is controversial and still limited to only few TLRs—TLR2, 3, 4, 5, and 9 [17, 27, 86, 98, 120, 126–128]. A recent advance in therapeutics has exploited both agonists and antagonists in a bid to develop an effective and long-lasting therapy for CRC [23, 51, 125]. Table 4 summarizes the different compounds which are being currently tested for the CRC therapy. While TLR agonists are used for their ability to activate the innate and adaptive immune responses, TLR antagonists are utilized for their immunosuppressive ability [86, 125, 129–131].
Table 4.
Summary of therapeutics targeting TLRs in CRC.
| Compound | Target TLRs | Drug | Clinical phase |
|---|---|---|---|
| BCG | TLR2/4 | Synthetic ssRNA | Phase I |
| MPL | TLR4 | Synthetic ssRNA | Phase I |
| CBLB502 | TLR5 | Flagellin | Phase I |
| Imiquimod | TLR7 | Small molecule ssRNA | Phase I/II/III |
| IMO2055 | TLR9 | CpG oligonucleotide | Phase I/II |
| MGN1703 | TLR9 | dSLIM | Phase II |
7. Conclusion
As inflammation is considered as the important risk factor for the CRC and TLRs playing a central role in inflammation-driven immune responses, TLRs have a universal role in carcinogenesis mechanism. Genetic polymorphisms in the important TLR genes especially TLR2 and TLR4 do have an effective modulatory bearing on the susceptibility to CRC, which is important in deducing them as one of the biomarkers for this malignancy.
Additionally, with the advent of new therapeutic strategies which target TLRs as legitimate targets of cancer therapy, the involvement of TLR SNPs and their risk modulation becomes ever more important to study and document. Numerous TLRs 2, 3, 4, 7, 8, and 9 have been identified as molecules of interest for the development of agonist, antagonist, and vaccines to develop a treatment strategy to control and prevent the development of CRC, which has a heavy bearing upon the TLR gene SNPs and the effects they have on immune modulation.
As this review was primarily focused on unraveling the importance of the TLR signaling and the SNPs thereof in CRC, the review is intentionally truncated with respect to the other aspects of TLR signaling. Additionally, we have discussed only those TLRs whose SNPs have been reported to be involved in CRC susceptibility. With the advancement of research on TLRs and the consequent unraveling of the role TLR SNPs play in CRC carcinogenesis, the prospects of the therapy targeting specific TLRs look promising. Currently, it would be safe to mention that combination therapy approach involving traditional drugs with the TLR agonists/antagonist may surely alleviate the benefits of treatment for CRC.
Acknowledgments
The authors do acknowledge their respective affiliations for the support.
Abbreviations
- ALRs:
AIM2-like receptors
- AP1:
Activator protein 1
- BCAP:
B-cell adaptor for phosphoinositide
- CLRs:
C-type lectin receptors
- CTD:
C-terminal domain
- DAMPs:
Damage/danger-associated molecular patterns
- DKs:
Dendritic cells
- IFN:
Interferon
- IKK:
IκB kinase
- IRAK:
IL-1R-associated kinase
- IRF-3:
Interferon regulatory factor-3
- LPS:
Lipopolysaccharide
- LRR:
Leucine-rich repeat
- MAMPs:
Microbe-associated molecular patterns
- MAPK:
Mitogen-activated protein kinases
- MyD88:
Myeloid differentiation primary response protein 88
- NEMO:
NF-κB essential modulator
- NF-κB:
Nuclear factor kappa B
- NKs:
Natural killer
- NLRs:
NOD-like receptors
- NOD:
Nucleotide-binding oligomerization domain
- NTD:
N-terminal domain
- PAMPs:
Pathogen-associated molecular patterns
- PRRs:
Pathogen recognition receptors
- RACK1:
Receptor for activated C kinase 1
- RIP1:
Receptor-interacting protein1
- RLRs:
Retinoic acid-inducible gene- (RIG-) I-like receptors
- SARM:
Sterile α- and armadillo-motif-containing protein
- TAB:
TAK1-binding protein 1
- TAK1:
TGF-β-activated kinase 1
- TANK:
TRAF family-member-associated NF-κB activator
- TBK1:
TANK-binding kinase 1
- TIR:
Toll-interleukin-1 receptor domain
- TIRAP:
TIR domain-containing adapter protein
- TLRs:
Toll-like receptors
- TRAF:
TNFR-associated factor
- TRAILR:
Tumor-necrosis factor-related apoptosis-inducing ligand receptor
- TRAM:
TRIF-related adaptor molecules
- TRIF:
TIR-domain-containing adaptor protein that induces IFN-γ
- TRIMs:
Tripartite motif-containing proteins
- TRIP:
TRAF-interacting protein
- XAMPs:
Xenobiotic-associated molecular patterns.
Data Availability
Data availability is not applicable, as it is a literature review.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
SSA conceptualized and designed the review, surveyed the existing literature, and wrote the manuscript. SN surveyed the existing literature, made the graphics, and edited the manuscript.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Ervik M., Lam F., et al. Cancer today (powered by GLOBOCAN 2018) IARC Cancer Base number. 2018:p. 15. June 2021, https://publications.iarc.fr/Databases/Iarc-Cancerbases/Cancer-Today-Powered-By-GLOBOCAN-2018. [Google Scholar]
- 3.Torre L. A., Islami F., Siegel R. L., Ward E. M., Jemal A. Global cancer in women: burden and trends. Cancer Epidemiology, Biomarkers & Prevention. 2017;26(4):444–457. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 4.Dekker E., Tanis P. J., Vleugels J. L. A., Kasi P. M., Wallace M. B. Colorectal cancer. The Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 5.Proenca M. A., de Oliveira J. G., Cadamuro A. C., et al. TLR2 and TLR4 polymorphisms influence mRNA and protein expression in colorectal cancer. World Journal of Gastroenterology. 2015;21(25):7730–7741. doi: 10.3748/wjg.v21.i25.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slattery M. L., Fitzpatrick F. A. Convergence of hormones, inflammation, and energy-related factors: a novel pathway of cancer etiology. Cancer Prevention Research (Phila) 2009;2(11):922–930. doi: 10.1158/1940-6207.CAPR-08-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wogan G. N., Dedon P. C., Tannenbaum S. R., Fox J. G. Infection, inflammation and colon carcinogenesis. Oncotarget. 2012;3:737–738. doi: 10.18632/oncotarget.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantovani A. Inflammation by remote control. Nature. 2005;435(7043):752–753. doi: 10.1038/435752a. [DOI] [PubMed] [Google Scholar]
- 9.Ullman T. A., Itzkowitz S. H. Intestinal inflammation and cancer. Gastroenterology. 2011;140(6):1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 10.Sameer A. S. Colorectal cancer: molecular mutations and polymorphisms. Frontiers in Oncology. 2013;3:p. 114. doi: 10.3389/fonc.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banday M. Z., Sameer A. S., Nissar S. Colorectal cancer and genetic polymorphism in key regulatory low penetrance genes. In: Sameer A. S., Banday M. Z., Nissar S., editors. Genetic Polymorphism and Cancer Susceptibility. Singapore: Springer; 2021. [DOI] [Google Scholar]
- 12.Vogelstein B., Fearon E. R., Hamilton S. R., et al. Genetic alterations during colorectal-tumor development. The New England Journal of Medicine. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 13.Armaghany T., Wilson J. D., Chu Q., Mills G. Genetic alterations in colorectal cancer. Gastrointestinal Cancer Research. 2012;5(1):19–27. [PMC free article] [PubMed] [Google Scholar]
- 14.Sameer A. S., Nissar S. Understanding epigenetics: an alternative mechanism of colorectal carcinogenesis. Current Colorectal Cancer Reports. 2016;12(3):113–122. doi: 10.1007/s11888-016-0317-6. [DOI] [Google Scholar]
- 15.Abreu M. T., Fukata M., Arditi M. TLR signaling in the gut in health and disease. Journal of Immunology. 2005;174(8):4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 16.Fukata M., Abreu M. T. Role of toll-like receptors in gastrointestinal malignancies. Oncogene. 2008;27(2):234–243. doi: 10.1038/sj.onc.1210908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slattery M. L., Herrick J. S., Bondurant K. L., Wolff R. K. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. International Journal of Cancer. 2012;130(12):2974–2980. doi: 10.1002/ijc.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Trejo-de la O A., Hernández-Sancén P., Maldonado-Bernal C. Relevance of single-nucleotide polymorphisms in human TLR genes to infectious and inflammatory diseases and cancer. Genes and Immunity. 2014;15(4):199–209. doi: 10.1038/gene.2014.10. [DOI] [PubMed] [Google Scholar]
- 20.West A. P., Koblansky A. A., Ghosh S. Recognition and signaling by toll-like receptors. Annual Review of Cell and Developmental Biology. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 21.El-Omar E., Ng M., Hold G. Polymorphisms in toll-like receptor genes and risk of cancer. Oncogene. 2008;27(2):244–252. doi: 10.1038/sj.onc.1210912. [DOI] [PubMed] [Google Scholar]
- 22.Kutikhin A. G. Association of polymorphisms in TLR genes and in genes of the toll-like receptor signaling pathway with cancer risk. Human Immunology. 2011;72(11):1095–1116. doi: 10.1016/j.humimm.2011.07.307. [DOI] [PubMed] [Google Scholar]
- 23.Li T. T., Ogino S., Qian Z. R. Toll-like receptor signaling in colorectal cancer: carcinogenesis to cancer therapy. World Journal of Gastroenterology. 2014;20(47):17699–17708. doi: 10.3748/wjg.v20.i47.17699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medvedev A. E. Toll-like receptor polymorphisms, inflammatory and infectious diseases, allergies, and cancer. Journal of Interferon & Cytokine Research. 2013;33(9):467–484. doi: 10.1089/jir.2012.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omrane I., Baroudi O., Kourda N., et al. Positive link between variant toll-like receptor 4 (Asp299Gly and Thr399Ile) and colorectal cancer patients with advanced stage and lymph node metastasis. Tumour Biology. 2014;35(1):545–551. doi: 10.1007/s13277-013-1075-6. [DOI] [PubMed] [Google Scholar]
- 26.Pandey S., Singh S., Anang V., Bhatt A. N., Natarajan K., Dwarakanath B. S. Pattern recognition receptors in cancer progression and metastasis. Cancer Growth Metastasis. 2015;8 doi: 10.4137/CGM.S24314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pimentel-Nunes P., Teixeira A. L., Pereira C., et al. Functional polymorphisms of toll-like receptors 2 and 4 alter the risk for colorectal carcinoma in Europeans. Digestive and Liver Disease. 2013;45(1):63–69. doi: 10.1016/j.dld.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Weber A. N., Försti A. Toll-like receptor genetic variants and colorectal cancer. Oncoimmunology. 2014;3(2, article e27763) doi: 10.4161/onci.27763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akira S., Takeda K., Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nature Immunology. 2001;2(8):675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 30.Ospelt C., Gay S. TLRs and chronic inflammation. The International Journal of Biochemistry & Cell Biology. 2010;42(4):495–505. doi: 10.1016/j.biocel.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Takeda K., Akira S. Toll-like receptors. Current Protocols in Immunology. 2015;109(1) doi: 10.1002/0471142735.im1412s109. [DOI] [PubMed] [Google Scholar]
- 32.Delneste Y., Beauvillain C., Jeannin P. Innate immunity: structure and function of TLRs. Medical Science (Paris) 2007;23(1):67–73. doi: 10.1051/medsci/20072316. [DOI] [PubMed] [Google Scholar]
- 33.Wang J. P., Bowen G. N., Padden C., et al. Toll-like receptor-mediated activation of neutrophils by influenza A virus. Blood. 2008;112(5):2028–2034. doi: 10.1182/blood-2008-01-132860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing J. J., Li M., Yuan Y. Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms in cancer: a meta-analysis. Gene. 2012;499(2):237–242. doi: 10.1016/j.gene.2012.03.045. [DOI] [PubMed] [Google Scholar]
- 35.Behzadi P., García-Perdomo H. A., Karpiński T. M. Toll-Like Receptors: General Molecular and Structural Biology. Journal of Immunology Research. 2021;2021 doi: 10.1155/2021/9914854.9914854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botos I., Segal D. M., Davies D. R. The structural biology of toll-like receptors. Structure. 2011;19(4):447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong E., Lee J. Y. Intrinsic and extrinsic regulation of innate immune receptors. Yonsei Medical Journal. 2011;52(3):379–392. doi: 10.3349/ymj.2011.52.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang J. Y., Lee J. O. Structural biology of the toll-like receptor family. Annual Review of Biochemistry. 2011;80:917–941. doi: 10.1146/annurev-biochem-052909-141507. [DOI] [PubMed] [Google Scholar]
- 39.Lee C. C., Avalos A. M., Ploegh H. L. Accessory molecules for toll-like receptors and their function. Nature Reviews. Immunology. 2012;12(3):168–179. doi: 10.1038/Nri3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brubaker S. W., Bonham K. S., Zanoni I., Kagan J. C. Innate immune pattern recognition: a cell biological perspective. Annual Review of Immunology. 2015;33(1):257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deretic V., Saitoh T., Akira S. Autophagy in infection, inflammation and immunity. Nature Reviews Immunology. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nature Immunology. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 43.Anderson K. V., Bokla L., Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the drosophila embryo: The induction of polarity by the Toll gene product. Cell. 1985;42(3):791–798. doi: 10.1016/0092-8674(85)90275-2. [DOI] [PubMed] [Google Scholar]
- 44.Schneider D. S., Hudson K. L., Lin T. Y., Anderson K. V. Dominant and recessive mutations define functional domains of toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes & Development. 1991;5(5):797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 45.Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A. The Dorsoventral Regulatory Gene Cassette sp atzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 46.El-Zayat S. R., Sibaii H., Mannaa F. A. Toll-like receptors activation, signaling, and targeting: an overview. Bulletin of the National Research Centre. 2019;43(1):p. 187. doi: 10.1186/s42269-019-0227-2. [DOI] [Google Scholar]
- 47.Lim K. H., Staudt L. M. Toll-like receptor signaling. Cold Spring Harbor Perspectives in Biology. 2013;5(1):p. a011247. doi: 10.1101/cshperspect.a011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie L., Cai S. Y., Shao J. Z., Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Frontiers in Immunology. 2018;9:p. 1523. doi: 10.3389/fimmu.2018.01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noreen M., Arshad M. Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility. Immunologic Research. 2015;62(2):234–252. doi: 10.1007/s12026-015-8640-6. [DOI] [PubMed] [Google Scholar]
- 50.Bell J. K., Mullen G. E., Leifer C. A., Mazzoni A., Davies D. R., Segal D. M. Leucine-rich repeats and pathogen recognition in toll-like receptors. Trends in Immunology. 2003;24(10):528–533. doi: 10.1016/S1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 51.Gao W., Xiong Y., Li Q., Yang H. Inhibition of toll-like receptor signaling as a promising therapy for inflammatory diseases: a journey from molecular to nano therapeutics. Frontiers in Physiology. 2017;8:p. 508. doi: 10.3389/fphys.2017.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang Z. L. Important aspects of toll-like receptors, ligands and their signaling pathways. Inflammation Research. 2010;59(10):791–808. doi: 10.1007/s00011-010-0208-2. [DOI] [PubMed] [Google Scholar]
- 53.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 54.Brennan J. J., Gilmore T. D. Evolutionary origins of toll-like receptor signaling. Molecular Biology and Evolution. 2018;35(7):1576–1587. doi: 10.1093/molbev/msy050. [DOI] [PubMed] [Google Scholar]
- 55.Leulier F., Lemaitre B. Toll-like receptors -- taking an evolutionary approach. Nature Reviews Genetics. 2008;9(3):165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 56.Reuven E. M., Fink A., Shai Y. Regulation of innate immune responses by transmembrane interactions: lessons from the TLR family. Biochimica et Biophysica Acta. 2014;1838(6):1586–1593. doi: 10.1016/j.bbamem.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 57.Fitzgerald K. A., Kagan J. C. Toll-like receptors and the control of immunity. Cell. 2020;180(6):1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Li J., Han J., Shu C., Xu T. Identification and characteristic analysis of TLR28: a novel member of the TLR1 family in teleost. Developmental & Comparative Immunology. 2016;62:102–107. doi: 10.1016/j.dci.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Medzhitov R., Preston-Hurlburt P., Janeway C. A., Jr. A human homologue of the _Drosophila_ Toll protein signals activation of adaptive immunity. Nature. 1997;388(6640):394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 60.Gay N. J., Symmons M. F., Gangloff M., Bryant C. E. Assembly and localization of toll-like receptor signalling complexes. Nature Reviews. Immunology. 2014;14(8):546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 61.Sellge G., Kufer T. A. PRR-signaling pathways: learning from microbial tactics. Seminars in Immunology. 2015;27(2):75–84. doi: 10.1016/j.smim.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 62.Gao D., Li W. Structures and recognition modes of toll-like receptors. Proteins. 2017;85(1):3–9. doi: 10.1002/prot.25179. [DOI] [PubMed] [Google Scholar]
- 63.Gay N. J., Gangloff M. Structure and function of toll receptors and their ligands. Annual Review of Biochemistry. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 64.Murad S. Toll-like receptor 4 in inflammation and angiogenesis: a double-edged sword. Frontiers in Immunology. 2014;5:p. 313. doi: 10.3389/fimmu.2014.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwandner R., Dziarski R., Wesche H., Rothe M., Kirschning C. J. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. The Journal of Biological Chemistry. 1999;274(25):17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 66.Underhill D. M., Ozinsky A., Smith K. D., Aderem A. Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(25):14459–14463. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 68.Poltorak A., He X., Smirnova I., et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 69.Hayashi F., Smith K. D., Ozinsky A., et al. The innate immune response to bacterial flagellin is mediated by toll-like receptor 5. Nature. 2001;410(6832):1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 70.Hemmi H., Kaisho T., Takeuchi O., et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nature Immunology. 2002;3(2):196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 71.Hemmi H., Takeuchi O., Kawai T., et al. A toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. Erratum in: Nature 2001 Feb 1; 409(6820):646. [DOI] [PubMed] [Google Scholar]
- 72.Akira S., Takeda K. Toll-like receptor signalling. Nature Reviews. Immunology. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 73.Leifer C. A., Medvedev A. E. Molecular mechanisms of regulation of toll-like receptor signaling. Journal of Leukocyte Biology. 2016;100(5):927–941. doi: 10.1189/jlb.2MR0316-117RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pradere J. P., Dapito D. H., Schwabe R. F. The yin and yang of toll-like receptors in cancer. Oncogene. 2014;33(27):3485–3495. doi: 10.1038/onc.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kirkwood J. M., Butterfield L. H., Tarhini A. A., Zarour H., Kalinski P., Ferrone S. Immunotherapy of cancer in 2012. CA: a Cancer Journal for Clinicians. 2012;62(5):309–335. doi: 10.3322/caac.20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diamond M. S., Kinder M., Matsushita H., et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. The Journal of Experimental Medicine. 2011;208(10):1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Palma M., Mazzieri R., Politi L. S., et al. Tumor-Targeted Interferon-α Delivery by Tie2-Expressing Monocytes Inhibits Tumor Growth and Metastasis. Cancer Cell. 2008;14(4):299–311. doi: 10.1016/j.ccr.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Pfeffer L. M. Biologic activities of natural and synthetic type I interferons. Seminars in Oncology. 1997;24:S9-63–S69-69. [PubMed] [Google Scholar]
- 79.Apte R. N., Voronov E. Is interleukin-1 a good or bad ‘guy’ in tumor immunobiology and immunotherapy? Immunological Reviews. 2008;222(1):222–241. doi: 10.1111/j.1600-065X.2008.00615.x. [DOI] [PubMed] [Google Scholar]
- 80.Balkwill F. Tumour necrosis factor and cancer. Nature Reviews. Cancer. 2009;9(5):361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 81.Grivennikov S. I., Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Annals of the Rheumatic Diseases. 2011;70(Supplement 1):i104–i108. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 82.Dutta J., Fan Y., Gupta N., Fan G., Gelinas C. Current insights into the regulation of programmed cell death by NF- κ B. Oncogene. 2006;25(51):6800–6816. doi: 10.1038/sj.onc.1209938. [DOI] [PubMed] [Google Scholar]
- 83.Wang C. Y., Mayo M. W., Korneluk R. G., Goeddel D. V., Baldwin A. S., Jr. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281(5383):1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 84.Wight T. N., Kang I., Evanko S. P., et al. Versican-a critical extracellular matrix regulator of immunity and inflammation. Frontiers in Immunology. 2020;11:p. 512. doi: 10.3389/fimmu.2020.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cui J., Chen Y., Wang H. Y., Wang R. F. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Human Vaccines & Immunotherapeutics. 2014;10(11):3270–3285. doi: 10.4161/21645515.2014.979640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X. X., Sun G. P., Meng J., et al. Role of toll-like receptor 4 in colorectal carcinogenesis: a meta-analysis. PLoS One. 2014;9(4, article e93904) doi: 10.1371/journal.pone.0093904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vidya M. K., Kumar V. G., Sejian V., Bagath M., Krishnan G., Bhatta R. Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. International Reviews of Immunology. 2018;37(1):20–36. doi: 10.1080/08830185.2017.1380200. [DOI] [PubMed] [Google Scholar]
- 88.Vijay K. Toll-like receptors in immunity and inflammatory diseases: past, present, and future. International Immunopharmacology. 2018;59:391–412. doi: 10.1016/j.intimp.2018.03.002. 2018 May 4. Erratum in: Int Immunopharmacol. 2018 Sep; 62:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mukherjee S., Huda S., Sinha Babu S. P. Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scandinavian Journal of Immunology. 2019;90(1, article e12771) doi: 10.1111/sji.12771. [DOI] [PubMed] [Google Scholar]
- 90.Joosten L. A., Abdollahi-Roodsaz S., Dinarello C. A., O'Neill L., Netea M. G. Toll-like receptors and chronic inflammation in rheumatic diseases: new developments. Nature Reviews Rheumatology. 2016;12(6):344–357. doi: 10.1038/nrrheum.2016.61. [DOI] [PubMed] [Google Scholar]
- 91.Li J., Wang X., Zhang F., Yin H. Toll-like receptors as therapeutic targets for autoimmune connective tissue diseases. Pharmacology & Therapeutics. 2013;138(3):441–451. doi: 10.1016/j.pharmthera.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thada S., Valluri V. L., Gaddam S. L. Influence of toll-like receptor gene polymorphisms to tuberculosis susceptibility in humans. Scandinavian Journal of Immunology. 2013;78(3):221–229. doi: 10.1111/sji.12066. [DOI] [PubMed] [Google Scholar]
- 93.Vaz J., Akbarshahi H., Andersson R. Controversial role of toll-like receptors in acute pancreatitis. World Journal of Gastroenterology. 2013;19(5):616–630. doi: 10.3748/wjg.v19.i5.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Skevaki C., Pararas M., Kostelidou K., Tsakris A., Routsias J. G. Single nucleotide polymorphisms of toll-like receptors and susceptibility to infectious diseases. Clinical and Experimental Immunology. 2015;180(2):165–177. doi: 10.1111/cei.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lucas C., Barnich N., Nguyen H. T. T. Microbiota, inflammation and colorectal cancer. International Journal of Molecular Sciences. 2017;18(6):p. 1310. doi: 10.3390/ijms18061310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lowe E. L., Crother T. R., Rabizadeh S., et al. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS One. 2010;5(9, article e13027) doi: 10.1371/journal.pone.0013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Messaritakis I., Stogiannitsi M., Koulouridi A., et al. Evaluation of the detection of toll-like receptors (TLRs) in cancer development and progression in patients with colorectal cancer. PLoS One. 2018;13(6, article e0197327) doi: 10.1371/journal.pone.0197327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nihon-Yanagi Y., Terai K., Murano T., Matsumoto T., Okazumi S. Tissue expression of toll-like receptors 2 and 4 in sporadic human colorectal cancer. Cancer Immunology, Immunotherapy. 2012;61(1):71–77. doi: 10.1007/s00262-011-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tang X. Y., Zhu Y. Q., Wei B., Wang H. Expression and functional research of TLR4 in human colon carcinoma. The American Journal of the Medical Sciences. 2010;339:319–326. doi: 10.1097/MAJ.0b013e3181cef1b7. [DOI] [PubMed] [Google Scholar]
- 100.Xu H., Wu Q., Dang S., et al. Alteration of CXCR7 expression mediated by TLR4 promotes tumor cell proliferation and migration in human colorectal carcinoma. PLoS One. 2011;6, article e27399 doi: 10.1371/journal.pone.0027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davoodi H., Seow H. F. Variant toll-like receptor4 (Asp299Gly and Thr399Ile alleles) and toll-like receptor2 (Arg753Gln and Arg677Trp alleles) in colorectal cancer. Iranian Journal of Allergy, Asthma, and Immunology. 2011;10(2):91–99. [PubMed] [Google Scholar]
- 102.Hosseini S., Mojtahedi Z., Beizavi Z., Khazraei H., Zamani M. Relationship between Arg753Gln toll-like receptor 2 and Asp299Gly toll-like receptor 4 genetic variations and susceptibility to colorectal cancer in southern Iran. Archives of Biological Sciences. 2018;70(4):775–779. doi: 10.2298/ABS180812043H. [DOI] [Google Scholar]
- 103.Sameer A. S., Banday M. Z., Nissar S. Mutations and Polymorphisms: What Is The Difference? In: Sameer A. S., Banday M. Z., Nissar S., editors. Genetic Polymorphism and cancer susceptibility. Singapore: Springer; 2021. [DOI] [Google Scholar]
- 104.TLR2. Gene Cards, The Human Database. 2021. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TLR2.
- 105.Zhu L., Yuan H., Jiang T., Wang R., Ma H., Zhang S. Association of TLR2 and TLR4 polymorphisms with risk of cancer: a meta-analysis. PLoS One. 2013;8(12, article e82858) doi: 10.1371/journal.pone.0082858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boraska Jelavić T., Barisić M., Drmic Hofman I., et al. Microsatelite GT polymorphism in the toll-like receptor 2 is associated with colorectal cancer. Clinical Genetics. 2006;70(2):156–160. doi: 10.1111/j.1399-0004.2006.00651.x. [DOI] [PubMed] [Google Scholar]
- 107.Yim J. J., Ding L., Schäffer A. A., Park G. Y., Shim Y. S., Holland S. M. A microsatellite polymorphism in intron 2 of human toll-like receptor 2 gene: functional implications and racial differences. FEMS Immunology and Medical Microbiology. 2004;40(2):163–169. doi: 10.1016/S0928-8244(03)00342-0. [DOI] [PubMed] [Google Scholar]
- 108.TLR3. Gene Cards, The Human Database. 2021. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TLR3.
- 109.Wang B. G., Yi D. H., Liu Y. F. TLR3 gene polymorphisms in cancer: a systematic review and meta-analysis. Chinese Journal of Cancer. 2015;34(6):272–284. doi: 10.1186/s40880-015-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Castro F. A., Försti A., Buch S., et al. TLR-3 polymorphism is an independent prognostic marker for stage II colorectal cancer. European Journal of Cancer. 2011;47(8):1203–1210. doi: 10.1016/j.ejca.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 111.TLR4. Gene Cards, The Human Database. 2021. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TLR4.
- 112.Nissar S., Sameer A. S., Rasool R., Qadri Q., Chowdri N. A., Rashid F. Role of _TLR4_ gene polymorphisms in the colorectal cancer risk modulation in ethnic Kashmiri population - A case-control study. Egyptian Journal of Medical Human Genetics. 2017;18(1):53–59. doi: 10.1016/j.ejmhg.2016.04.004. [DOI] [Google Scholar]
- 113.Sheng W. Y., Yong Z., Yun Z., Hong H., Hai L. L. Toll-like receptor 4 gene polymorphisms and susceptibility to colorectal cancer: a meta-analysis and review. Archives of Medical Science. 2015;11(4):699–707. doi: 10.5114/aoms.2015.53288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Arbour N. C., Lorenz E., Schutte B. C., et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nature Genetics. 2000;25(2):187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 115.Moaaz M., Youssry S., Moaz A., Abdelrahman M. Study of toll-like receptor 4 gene polymorphisms in colorectal cancer: correlation with clinicopathological features. Immunological Investigations. 2020;49(5):571–584. doi: 10.1080/08820139.2020.1716787. [DOI] [PubMed] [Google Scholar]
- 116.Kopp T. I., Vogel U., Tjonneland A., Andersen V. Meat and fiber intake and interaction with pattern recognition receptors (TLR1, TLR2, TLR4, and TLR10) in relation to colorectal cancer in a Danish prospective, case-cohort study. The American Journal of Clinical Nutrition. 2018;107(3):465–479. doi: 10.1093/ajcn/nqx011. [DOI] [PubMed] [Google Scholar]
- 117.TLR5. Gene Cards, The Human Database. 2021. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TLR5.
- 118.Klimosch S. N., Försti A., Eckert J., et al. Functional TLR5 genetic variants affect human colorectal cancer survival. Cancer Research. 2013;73(24):7232–7242. doi: 10.1158/0008-5472.CAN-13-1746. [DOI] [PubMed] [Google Scholar]
- 119.TLR9. Gene Cards, The Human Database. 2021. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TLR3.
- 120.Fűri I., Sipos F., Germann T. M., et al. Epithelial toll-like receptor 9 signaling in colorectal inflammation and cancer: clinico-pathogenic aspects. World Journal of Gastroenterology. 2013;19(26):4119–4126. doi: 10.3748/wjg.v19.i26.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao C., Qiao T., Zhang B., Yuan S., Zhuang X., Luo Y. TLR9 signaling activation at different stages in colorectal cancer and NF-kappaB expression. Oncotargets and Therapy. 2018;11:5963–5971. doi: 10.2147/OTT.S174274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Braunstein M. J., Kucharczyk J., Adams S. Targeting toll-like receptors for cancer therapy. Targeted Oncology. 2018;13(5):583–598. doi: 10.1007/s11523-018-0589-7. [DOI] [PubMed] [Google Scholar]
- 123.Brunner R., Jensen-Jarolim E., Pali-Schöll I. The ABC of clinical and experimental adjuvants--a brief overview. Immunology Letters. 2010;128(1):29–35. doi: 10.1016/j.imlet.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kaczanowska S., Joseph A. M., Davila E. TLR agonists: our best frenemy in cancer immunotherapy. Journal of Leukocyte Biology. 2013;93(6):847–863. doi: 10.1189/jlb.1012501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moossavi S., Rezaei N. Toll-like receptor signalling and their therapeutic targeting in colorectal cancer. International Immunopharmacology. 2013;16(2):199–209. doi: 10.1016/j.intimp.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 126.Bednarczyk M., Muc-Wierzgoń M., Walkiewicz K., Kokot T., Fatyga E., Mazurek U. Profile of gene expression of TLR-signaling pathways in colorectal cancer tissues. International Journal of Immunopathology and Pharmacology. 2017;30(3):322–326. doi: 10.1177/0394632017716048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang E. L., Qian Z. R., Nakasono M., et al. High expression of toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. British Journal of Cancer. 2010;102(5):908–915. doi: 10.1038/sj.bjc.6605558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoshida T., Miura T., Matsumiya T., et al. Toll-like receptor 3 as a recurrence risk factor and a potential molecular therapeutic target in colorectal cancer. Clinical and Experimental Gastroenterology. 2020;13:427–438. doi: 10.2147/CEG.S252157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gambara G., De Cesaris P., De Nunzio C., et al. Toll-like receptors in prostate infection and cancer between bench and bedside. Journal of Cellular and Molecular Medicine. 2013;17(6):713–722. doi: 10.1111/jcmm.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hennessy E. J., Parker A. E., O'Neill L. A. Targeting toll-like receptors: emerging therapeutics? Nature Reviews. Drug Discovery. 2010;9(4):293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- 131.Vacchelli E., Eggermont A., Sautès-Fridman C., et al. Trial watch: toll-like receptor agonists for cancer therapy. Oncoimmunology. 2013;2(8, article e25238) doi: 10.4161/onci.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability is not applicable, as it is a literature review.


