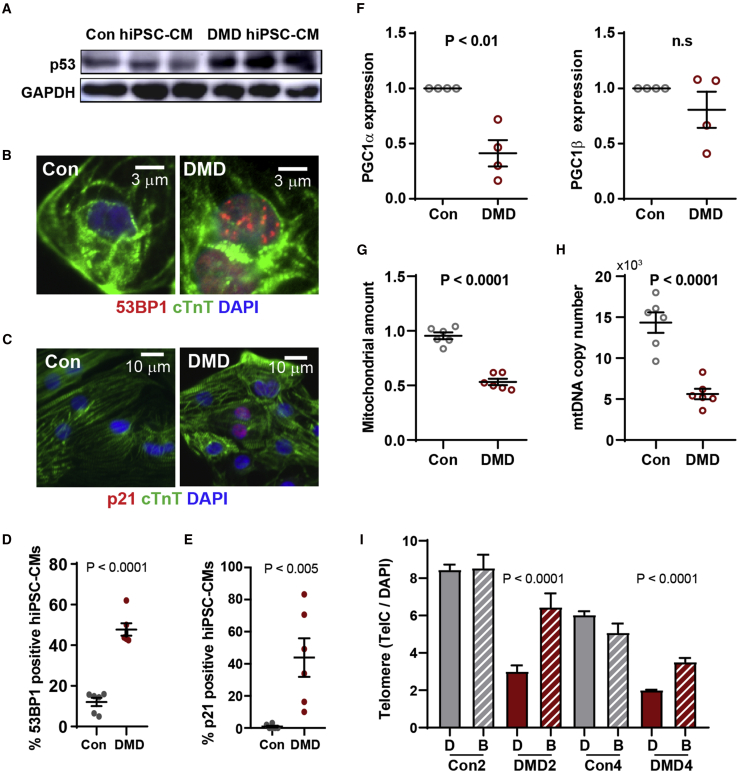
Figure 5.
DMD hiPSC-CMs exhibit p53 upregulation
(A–E) Representative micrographs of (A) p53 activation by immunoblotting (n = 3 independent experiments), (B) DNA damage 53BP1 foci by immunofluorescence (n = 6 independent experiments), and (C) p21 (n = 6 independent experiments) in cardiac troponin T+ hiPSC-CMs are shown and (D and E) quantified, respectively.
(F) Reduced expression levels of PGC-1α, master regulator of mitochondrial biogenesis, determined by qRT-PCR (n = 6 independent experiments).
(G and H) (G) Mitochondria amount (n = 6 independent experiments) and (H) mitochondrial copy number (n = 6 independent experiments) in hiPSC-CMs were assessed by MitoTracker Green and qRT-PCR using mitochondrial gene (Nd2) to nuclear DNA (Nrf1) primers, respectively. Data represent mean ± SEM. Student's t was test used for statistical analysis.
(I) Telomere loss prevented when contraction of DMD hiPSC-CMs was inhibited between days 20 and 30 using blebbistatin was quantified (n = 3 independent experiments, 35–1,834 cells analyzed). Data represent mean ± SEM. Student's t test was used for statistical analysis.