FIGURE 5.
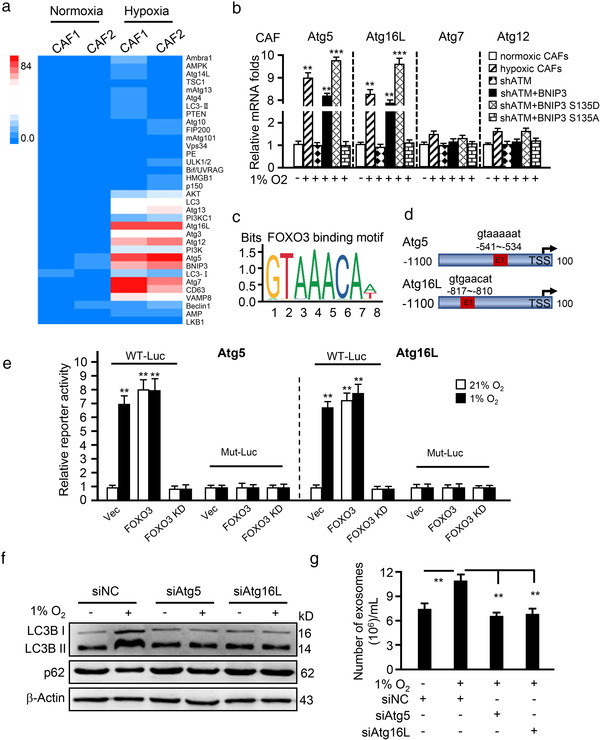
Phosphorylated BNIP3 upregulates Atg5 and Atg16L to induce autophagy and exosome release. (a) Geometric mean‐centred, hierarchical cluster heat‐map from microarray data of normoxic CAFs and hypoxic CAFs, and targets of interest are shown. (b) qRT‐PCR to test Atg5, Atg16L, Atg7 and Atg12 expression in normoxic CAFs, hypoxic CAFs, endogenous ATM‐silenced CAFs (shATM), and endogenous ATM‐silenced CAFs transfected with ectopic wild type BNIP3 (shATM+BNIP3) or BNIP3 mutant (S135D mutant, shATM+BNIP3 S135D; or S135A mutant, shATM+BNIP3 S135A). (c, d) Schematic diagram shows the FOXO3 binding motif (c) and binding sites in ATG5 and ATG16L promoter (d). (e) The Atg5 or Atg16L wild type LUC reporter (with FOXO3 binding site, WT‐Luc) or mutant LUC reporter (FOXO3 binding site being mutated, Mut‐Luc) were transfected into CAFs (control CAFs), engineered CAFs with or without FOXO3 and cultured at indicated condition for 30 h, the luciferase activities of Atg5 and Atg16L were measured. **P < 0.01: compared with vector (Vec) group in normoxia (21%). (f) Western blotting to determine autophagy protein markers LC3B II and p62 in normoxia and hypoxic CAFs with or without silenced‐Atg5 or Atg16L. (g) Concentration of exosomes isolated from normoxic or hypoxic CAFs with silenced‐Atg5 or Atg16L was evaluated by NTA assay. (**P < 0.01; ***P < 0.001.)
