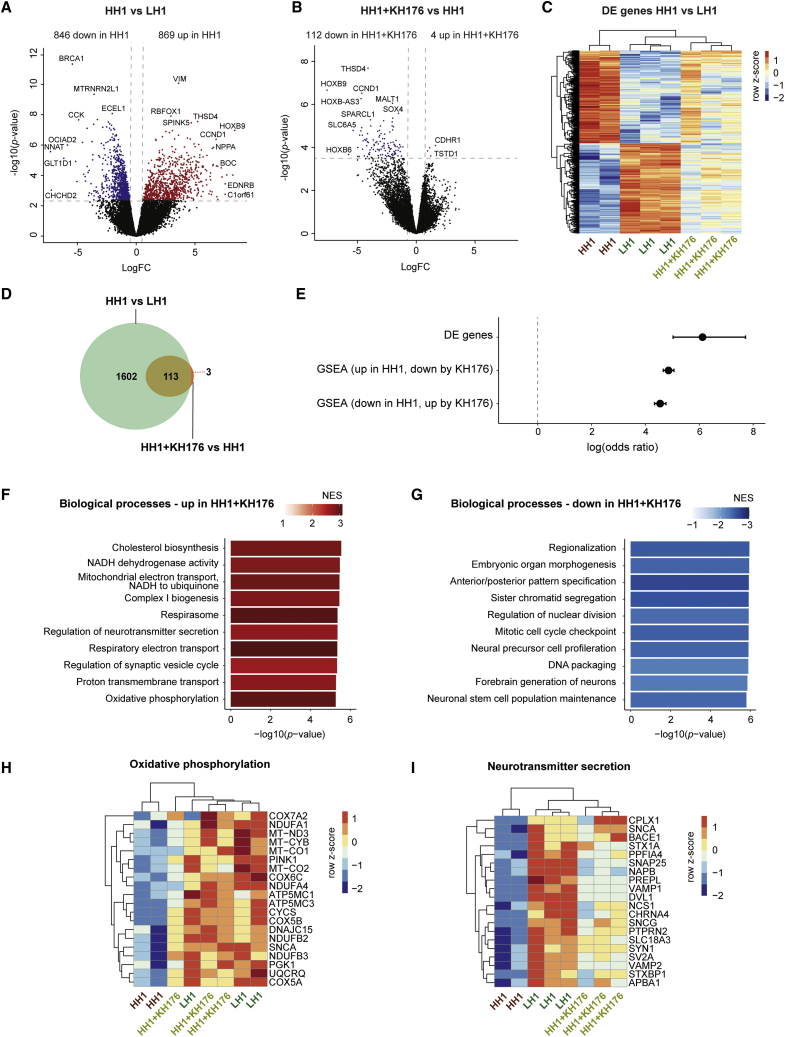
Figure 6.
Effects of sonlicromanol (KH176) treatment on gene expression in HH iNeurons for isogenic set 1 (HH1)
(A and B) Volcano plot showing DE genes in (A) HH1 compared with LH1 iNeurons or (B) HH1+KH176 compared with untreated HH1 iNeurons (genes with adjusted p < 0.05 are labeled), with upregulated genes in red (logFC > 0) and downregulated genes in blue (logFC<0).
(C) Heatmap showing expression of DE genes in HH1 versus LH1, for LH1 (n = 3), HH1 (n = 2), and HH1+KH176 (n = 3) samples. Voom-transformed and batch-corrected counts per million (log2 scale) were scaled per gene.
(D) Venn diagram showing overlap between DE genes in HH1 versus LH1, and DE genes in HH1+KH176 versus HH1.
(E) The log(odds ratio) is shown for overlap between DE genes in HH1 versus LH1 and HH1+KH176 versus HH1, and for overlap between gene sets enriched for upregulated genes in HH1 versus LH1 and gene sets enriched for downregulated genes in HH1+KH176 versus LH1, and vice versa. Error bars represent the 95% confidence interval.
(F and G) Bar plot showing 10 gene sets that are among the top gene sets enriched for (F) upregulated or (G) downregulated genes in HH1+KH176 versus HH1. The −log10(p value) (x axis) and the NES (color coded: red for positive NES, blue for negative NES) for Reactome pathways and GO terms representing BPs are shown.
(H and I) Heatmap showing top 20 leading edge genes in HH1+KH176 versus HH1 for the gene sets (H) oxidative phosphorylation, and (I) neurotransmitter secretion, for LH1, HH1, and HH1+KH176 samples. Voom-transformed and batch-corrected counts were scaled per gene.
See also Figures S2+ S6.