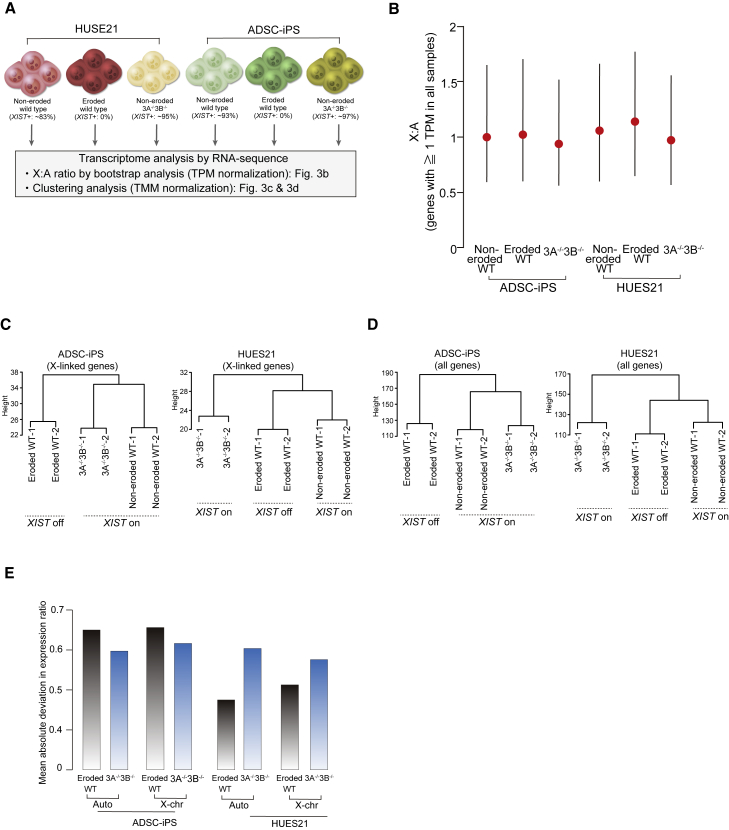
Figure 3.
Influence of DNMT3A and DNMT3B double mutations on transcriptional status
(A) Experimental scheme for RNA-sequence analysis. Non-eroded (wild type), eroded (wild type), and DNMT3A−/−3B−/− in the HUES21 and ADSC-iPS lines were used for the assay. In each line, two replicates were analyzed.
(B) Bootstrapped X:A ratios in each cell line. Genes with ≥1 TPM in all cell lines were used for the assay. X:A ratios with 95% confidence intervals are shown and the mean ratio is indicated by a red circle.
(C and D) Hierarchical clustering analysis using X-linked genes (C) and all genes (D). TMM normalization was employed and genes with >1 TMM in each cell line were used.
(E) Bar plot illustrating the mean absolute deviation in expression ratios of eroded or DNMT3A−/−3B−/− cell lines compared with non-eroded wild-type cell lines. The genes used in the hierarchical clustering analysis were used for the calculation.