Figure 4.
Downregulation of ER stress pathways by hypoxia during DXI-mediated expansion of human LTR-HSCs
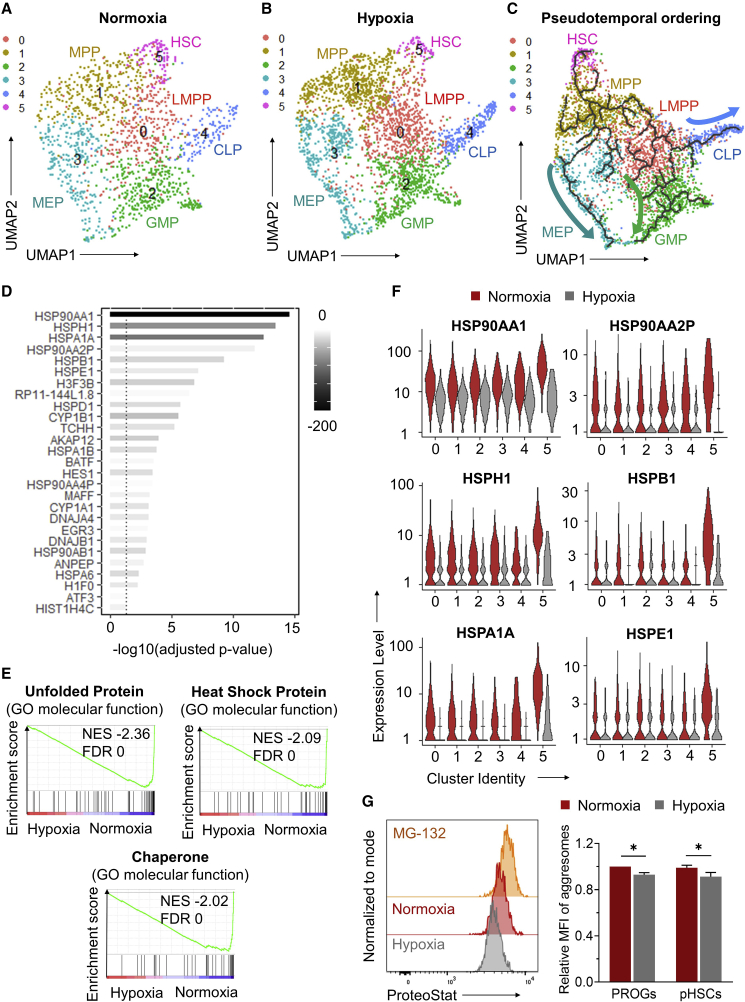
(A and B) UMAP visualization of scRNA-seq data for CD34+ cells treated in vessels coated with optimized densities of DXI under normoxic (A) or hypoxic (B) conditions. A total of six cell clusters were identified and annotated as lymphoid-primed multipotent progenitors (LMPP, cluster 0), multipotent progenitors (MPP, cluster 1), granulocyte-monocyte progenitors (GMP, cluster 2), megakaryocytic-erythroid progenitors (MEP, cluster 3), common lymphoid progenitors (CLP, cluster 4), and hematopoietic stem cells (HSC, cluster 5).
(C) Reconstruction of the hematopoietic hierarchy pseudotime ordering by Monocle in the integrated dataset.
(D) List of 27 differentially expressed genes (DEGs) (all downregulated) identified by comparing the transcriptome of HSC cluster 5 from normoxic and hypoxic cultures. These genes represent the overlap between two lists of DEGs identified using two independent statistical methods, Wilcoxon rank sum (Figure S4C) and DESeq2 (Figure S4D).
(E) Enrichment plots showing the relative expression of genes associated with unfolded protein, heat-shock protein, and chaperone pathways. NES, normalized enrichment score; FDR, false discovery rate.
(F) Violin plots showing expression levels of the top downregulated genes identified in (D).
(G) Quantification of protein aggregates (aggresomes) in human CD34+CD38+ progenitors (PROGs) and phenotypically defined CD34+CD38−CD90+CD45RA− LTR-HSC populations (pHSCs) cultured with optimized DXI densities under normoxic or hypoxic conditions, as measured using a ProteoStat staining approach. Left: representative flow-cytometry histograms; the proteasome inhibitor MG-132 was used as positive control to define a population of ProteoStathigh population. Right: summary of relative mean fluorescence intensity (MFI) of aggresomes (n = 9 technical replicates, from three independent donors). Data are displayed as mean ± SEM. One-way repeated-measures ANOVA was used. ∗p ≤ 0.05.
See also Figures S4 and S5; Tables S3, S4, and S5.