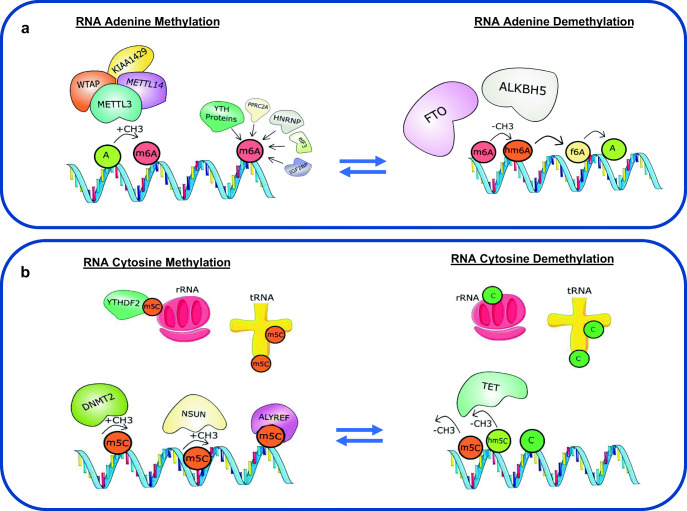
Fig. 2.
Dynamic regulation of adenine and cytosine RNA methylation and demethylation. a Adenine residues (A) are methylated (m6A) under enzymatic action of the methyltransferase complex comprising of methyltransferase like 3 (METTL3), METTL14, Wilms’ tumour–associated protein 1 (WTAP1) and KIAA1429. Proteins such as the YT521-B homology (YTH) family of proteins, heterogeneous nuclear ribonucleoprotein (HNRNP) protein, eukaryotic initiation factor 3 (eIF3), proline rich coiled-coil 2A (PPRC2A), insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) act as readers that recognize and bind to m6A. Conversely, m6A can also be removed through enzymatic demethylation by oxidation by both fat mass and obesity–associated protein (FTO) and alkylation repair homologue 5 (ALKBH5) to N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A). m6A plays a critical role in regulating mRNA fate, including pre-mRNA splicing, mRNA stability, nuclear transport and translation. b Cytosine bases (C) in RNA can also become methylated by the methyltransferase action of the NOL1/NOP2/Sun (NSUN) domain-containing family and the DNA methyltransferase 2 enzyme (DNMT2), leading to formation of 5-methylcytosine (m5C) on different RNA species including messenger RNA, ribosomal RNA (rRNA) and transfer RNA (tRNA). Proteins such as ALYREF and YTHDF2 act as m5C reader proteins and recognize and bind to m5C on RNA. m5C marks can be removed via demethylation by the action of the ten-eleven translocation (TET) enzymes leading to the formation of hydroxy-m5C (hm5C). RNA cytosine methylation has been shown to influence numerous aspects of RNA biology, including structure, stability and translation of mRNA along with the biogenesis and function of ribosomes