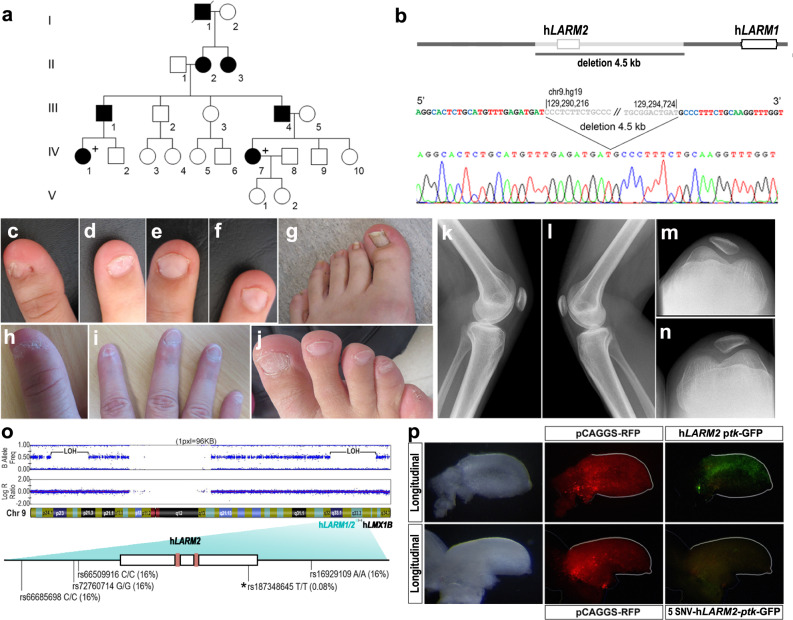
Fig. 5. Clinical features of LARM loss-of-function.
a Pedigree of a family with a LARM deletion. b The 4.5 kb region deleted removes all of LARM2. c–g, k–n Phenotypic images of individual IV-7. c–d Koïlonychia of thumb and 2nd finger. e–f Triangular lunulae of 3rd and 4th fingers. g Nail dysplasia of the hallux showing longitudinal striations. h–j Phenotypic images of individual IV-1. h Koïlonychia of thumb. i Hypoplastic nails, ungueal dysplasia of 2nd finger. j Ungueal dysplasia of right foot predominating on 1st and 5th toes. k–n Knee X-rays showing bilateral hypoplasia of the patella. o Schematic of the patient’s chromosome 9 showing large segments of the chromosome with loss of heterozygosity (LOH), i.e., homozygosity, when comparing the allele frequencies to the Log R ratio of the alleles. One of the homozygous regions includes the LARM-LMX1B locus. The homozygous hLARM2 sequence showing the five single nucleotide variations (SNVs). The asterisk indicates the rare (0.08%) sequence in LARM2. The LMX1B binding sites are indicated as clay-red boxes. p Using site-directed mutagenesis, we generated a human LARM2 construct containing the patient´s 5 SNVs; following electroporation into embryonic chick wings, the patient-LARM2 sequence showed markedly reduced activity (n = 6; compare with the activity of the common hLARM2 sequence in Fig. 4a). Interestingly, mutation of only the single SNV within the conserved LARM2 region did not alter LARM2 activity.