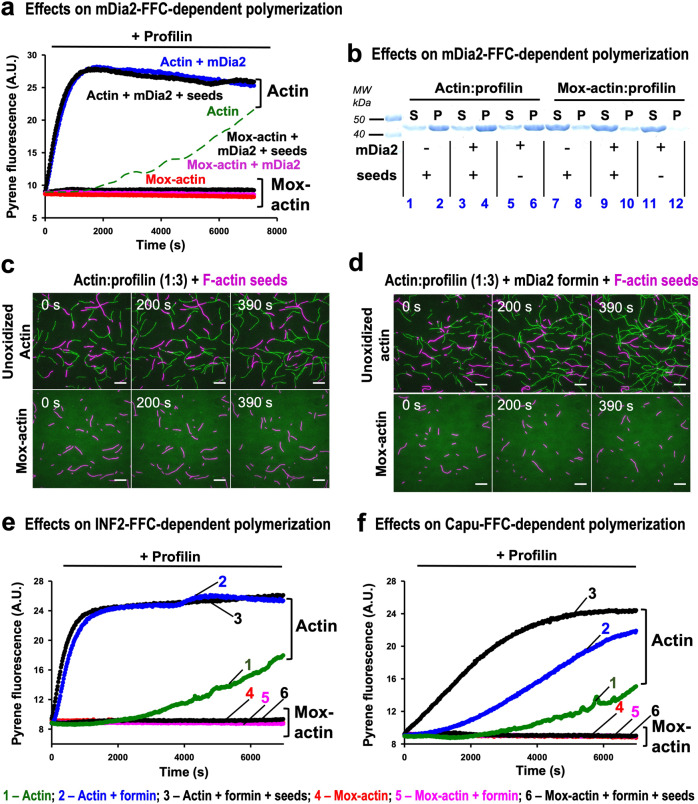
Fig. 2. Mox-actin–profilin complexes markedly inhibit formin-dependent actin polymerization.
a–d Assembly of unoxidized and Mox-actin in the presence of profilin and mDia2-FFC formin construct (with or without phalloidin-stabilized F-actin seeds). Results obtained by three different methods—pyrene fluorescence measurements (a), high-speed co-sedimentation (b), and TIRF microscopy (c, d)—are consistent with the absence of mDia2-FFC- -driven Mox-actin assembly in the presence of profilin. n = 2 separate experiments with similar results for each of a–d. a, b [Actin] =3 µM; [profilin] = 9 µM; [mDia2-FFC] = 30 nM; [F-actin-phalloidin seeds] = 0.25 µM (stabilized with phalloidin (Ph) at 1:50 Ph:actin molar ratio). A.U. arbitrary units. c, d [Actin] = 0.5 µM, [Mox-actin] = 1.4 µM (0.4 µM above each of their critical concentrations); Profilin-to-actin ratios = 3:1. [mDia2-FFC] = 1.5 nM. Note that the profilin–actin control sample in (a green trace) is a smoothed fit through the data points and is therefore shown as a dashed line (see Source Data file). This condition is repeated in the green traces in Figs. 1d, 2e, 2f, and Supplemental Fig. 2. Scale bars (c, d) = 10 µm. e, f INF2 (e) and Drosophila Capu (f) formins do not support Mox-actin assembly in the presence of profilin. Traces obtained are labeled as follows (similar to Fig. 2a): 1—Actin; 2—Actin + formin; 3—Actin + formin + seeds; 4—Mox-actin; 5—Mox-actin + formin; 6—Mox-actin + formin + seeds. n = 2 separate experiments with similar results for each of e and f. [Actin] = 3 µM; [profilin] = 9 µM; [INF2-FFC] = 10 nM; [Capu-FFC] = 20 nM; [F-actin-phalloidin seeds] = 0.25 µM (stabilized with phalloidin (Ph) at 1:50 Ph:actin molar ratio). Note that formins have different potencies in promoting actin assembly (e.g., refs. 78,79). Therefore, different concentrations of formins’ were used in the reactions. Source data for Fig. 2 including uncropped gels are provided as a Source Data file.