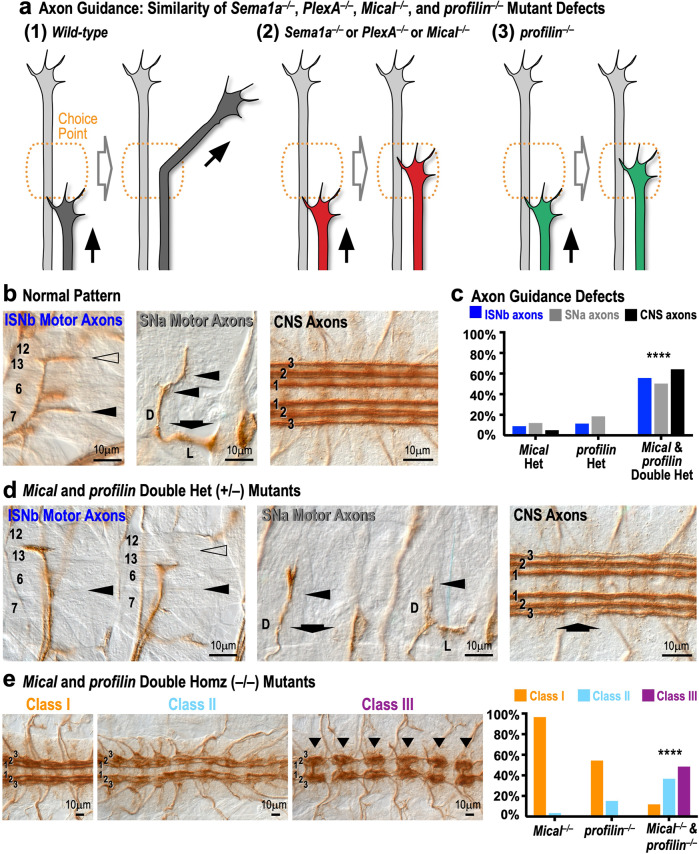
Fig. 5. Mical and profilin functionally interact in vivo to guide axons.
a Similarity of Sema1a−/−, PlexA−/−, Mical−/−, and profilin−/− mutant axon guidance defects. (1) Axons elongate/fasciculate with other axons (arrow, left). At choice points (orange), axons remodel themselves to grow in new directions (arrow, right). (2) Loss of Sema1a, PlexA, or Mical results in axons (red) that stall/don’t respond at choice points and thus do not change their trajectory. (3) Loss of profilin also results in axons (green) that stall at choice points and don’t change their trajectory. b–e Mical and profilin combine to guide axons in vivo. b Normal pattern of motor and CNS axon guidance. ISNb motor axons: defasciculate/separate from their main nerve and project to muscles 6/7 (filled arrowhead) and muscles 12/13 (open arrowhead). SNa motor axons: defasciculate to give rise to a dorsal (D) and lateral (L) branch (arrow)—and dorsal branch axons then make two characteristic turns (filled arrowheads). CNS axons: project within three 1D4-positive longitudinal bundles (1, 2, 3). c, d Compared to either heterozygote (Het) alone, Mical+/– and profilin+/– double Hets give rise to significant guidance defects including: absent/abnormal innervation of muscles 6/7 (ISNb, closed arrowhead) and 12/13 (ISNb, open arrowhead), absence of SNa lateral branch (SNa, arrow), failure of SNa axons to turn (SNa, arrowheads), and decreases in axons in the third CNS longitudinal bundle (CNS, arrowhead). Mical Het = MicalI1367/+ (ISNb and SNa, n = 100 hemisegments assessed across 10 animals; CNS, n = 20 animals). profilin Het = chic221/+ (ISNb and SNa, n = 70 hemisegments assessed across 7 animals; CNS, n = 20 animals). Mical & profilin Double Het = MicalI1367/+ & chic221/+ (ISNb and SNa, n = 140 hemisegments assessed across 14 animals; CNS, n = 14 animals). ****p < 0.0001; Χ2 test. e Mical−/− and profilin−/− double homozygous (Homz) mutants exhibit significantly more severe guidance defects than either homozygous mutant alone—including Class III defects, in which axons are excessively/repetitively bundled together (arrowheads). Scoring system adapted from41. Defects: Class I (affecting longitudinal bundle 3), Class II (affecting longitudinal bundles 2 and 3), Class III (affecting longitudinal bundles 1, 2, and 3). Mical−/− = MicalDf(3R)swp2/MicalDf(3R)swp2 (n = 29 animals; see also9,40), profilin−/− = chic221/chic221 (n = 46 animals; see also41,51,52), Mical−/− & profilin−/− = MicalDf(3R)swp2/MicalDf(3R)swp2 & chic221/chic221 (n = 109 animals). ****p < 0.0001; Χ2 test. b–e ≥2 independent experiments gave similar results. Source for Fig. 5 are provided as a Source Data file.