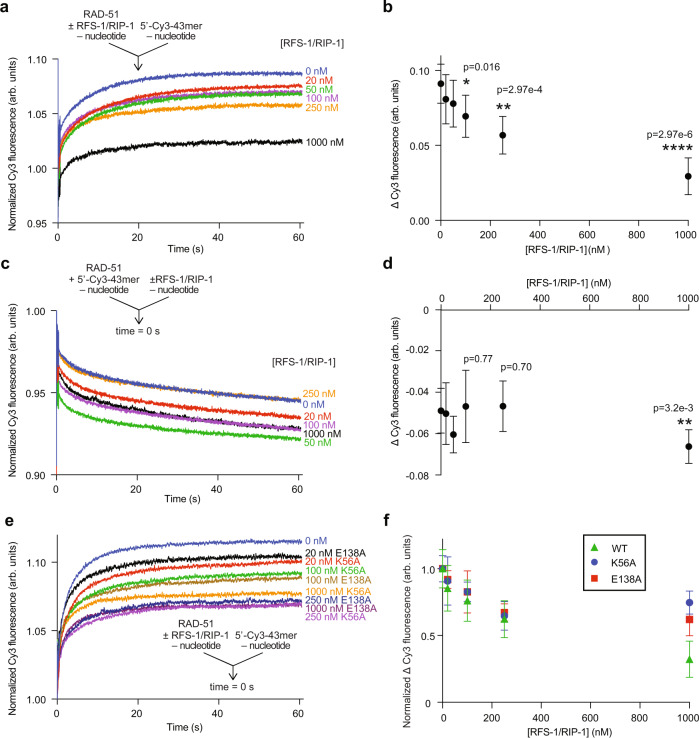
Fig. 2. RFS-1/RIP-1 inhibits the binding of RAD-51 to ssDNA in the absence of nucleotide cofactors.
Average normalized Cy3-43mer fluorescence profiles plotted as a function of time. The arrow indicates the components of the two syringes rapidly mixed at the 0 s time point in a stopped flow instrument. RAD-51 (1 μM) was either pre-mixed with increasing amount of RFS-1/RIP-1 and then was mixed with 15 nM Cy3-43mer ssDNA (n = 6–7) (a) or RAD-51 was pre-mixed with ssDNA and afterwards mixed with RFS-1/RIP-1 in the absence of nucleotide and analyzed in stopped flow machine (n = 7–8) (c). Corresponding evaluations of average Δ Cy3 fluorescence as a function of concentration of RFS-1/RIP-1 for the experiments in (a and c) are presented in (b and d) respectively (mean; errors: s.d.). p values are relative to RAD-51 alone data obtained by Student’s t test (two-tailed): *p < 0.05; **p < 0.01; ****p < 0.0001. e Effect of Walker box mutants of RFS-1 (K56A and E138A) on RAD-51 in the absence of nucleotide. The experiment was done similarly as in (a) (n = 5–8). f Normalized Δ Cy3 fluorescence relative to RAD-51 alone at different RFS-1/RIP-1 mutant concentrations from data shown in (e). Data for wild-type RAD-51 are taken from (b) for comparison (mean, errors: s.d.). Source data are provided as a Source Data file.