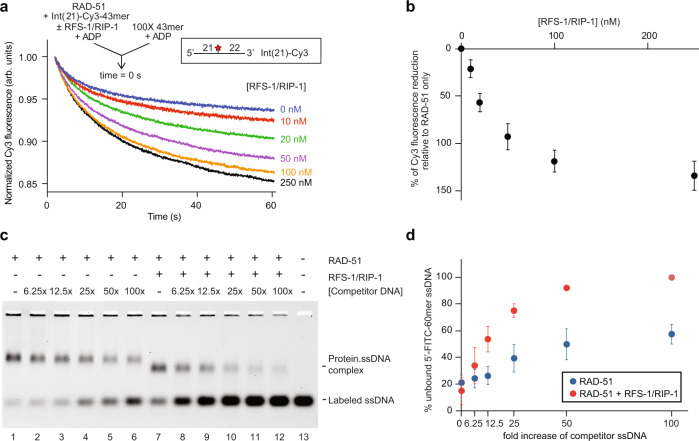
Fig. 3. RFS-1/RIP-1 destabilizes RAD-51-ssDNA filaments in the presence of ADP.
a Average normalized Cy3-43mer fluorescence profiles plotted as a function of time. The arrow indicates the components of the two syringes rapidly mixed at the 0 s time point in a stopped flow instrument. RAD-51-ssDNA filaments pre-formed with RAD-51 (1 μM) and Int(21)-Cy3 labelled 43mer ssDNA (15 nM) and indicated concentrations of RFS-1/RIP-1 were mixed for 10 min in the presence of ADP. The mixture was then challenged with 100-fold excess of unlabelled 43mer and analyzed (n = 5–8). b Graph of RFS-1/RIP-1 concentration-dependence change of Cy3 fluorescence from (a) (mean; errors: s.d.). c Representative EMSA gel (n = 4) demonstrating destabilization of RAD-51-ssDNA filaments in the presence of ADP. RAD-51 (1 μM) and RFS-1/RIP-1 (0.5 μM) were pre-incubated before addition of 5′-FITC-61mer ssDNA (10 nM) for 10 min and then challenged with increasing amounts of unlabelled 61mer ssDNA for a further 10 min. Protein-DNA complexes were crosslinked and resolved in agarose gels. d The amount of unbound 5′-FITC-61mer ssDNA from (c) (n = 4) was quantified. The average percentage of unbound 5′-FITC-61mer ssDNA was plotted as a function of the relative excess concentration of unlabelled 61mer over 5′-FITC-61mer ssDNA (mean; errors: s.d.). Source data are provided as a Source Data file.