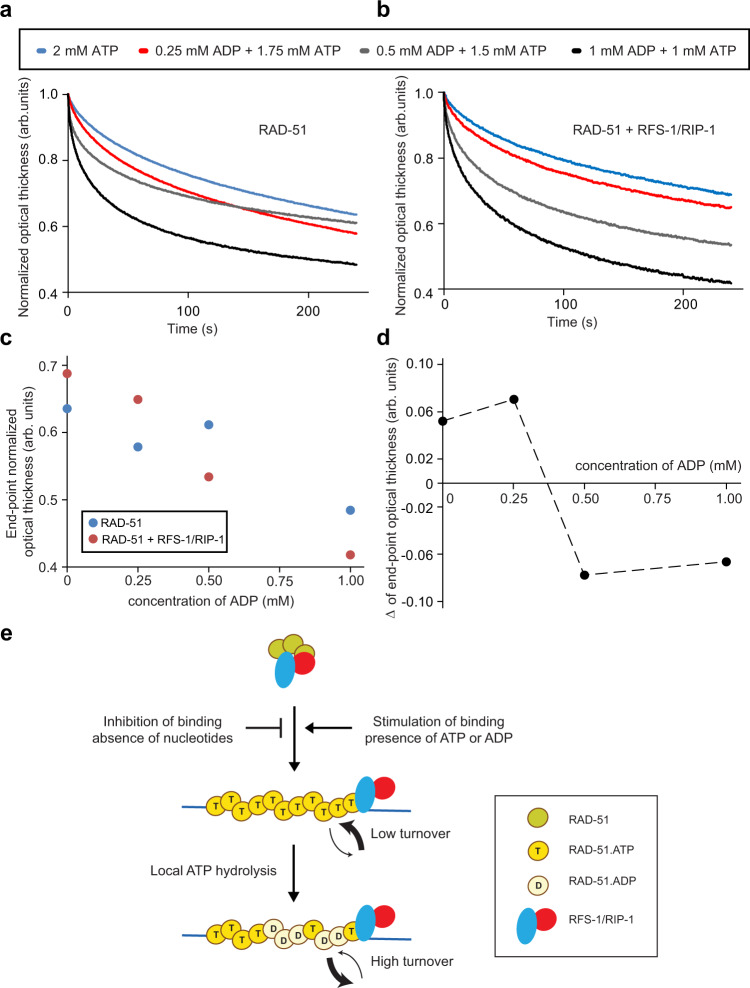
Fig. 5. A threshold of ADP in the RAD-51-ssDNA filament triggers a switch in RFS-1/RIP-1 activity from stabilization to destabilization.
a, b Normalized dissociation phases obtained from BLI experiments using 3′ biotinylated ssDNA (15 nM) where RAD-51 (1 μM) alone (a) or in the presence of RFS-1/RIP-1 (0.25 μM) (b) was loaded on the biosensor together with different ATP/ADP ratios. The biosensor with bound proteins was then submerged in buffer without any nucleotide cofactor and 1000-fold excess of unlabelled ssDNA to facilitate dissociation. c The corresponding endpoint in optical thickness across the dissociation phase for the data presented in (a and b) is plotted as a function of ADP concentration. d The difference in the values in (c) for RAD-51 ±RFS-1/RIP-1 was plotted to estimate the break point where stabilization switches to destabilization. e Schematic model of the action of RAD51 paralogs based on our results. RAD-51 binding to ssDNA is inhibited by RAD51 paralogs in the absence of nucleotides, whereas in ATP or ADP the formation of nucleofilaments is stimulated. Filaments undergoing local ATP hydrolysis are destabilized leading to RFS-1/RIP-1-mediated filament turnover. Source data are provided as a Source Data file.