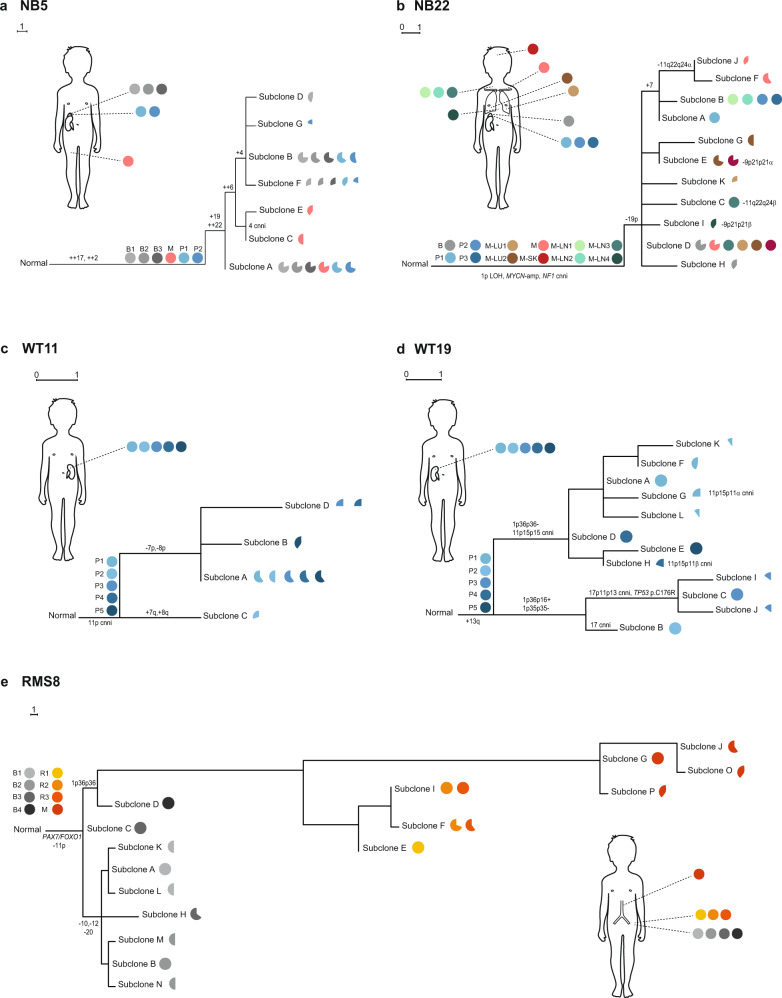
Fig. 2. Phylogenetic trees of childhood cancers.
At the stem of the maximum parsimony trees illustrated here, the biopsies available from each patient are denoted. The genetic alterations belonging to the stem are present in all cells in all samples, indicated by filled pies. The endpoints represent cell populations harboring distinct genomic profiles (subclones), whose fractions per sample are visualized by pie charts. The scale bar indicates the distance corresponding to one genetic aberration. Gains and losses of chromosomes or segments of chromosomes that are characteristic of each tumor subtype are indicated by + and – signs. a In NB5 samples are available from the primary tumor before treatment (B1–B3), a synchronous metastasis (M), and the primary tumor post treatment (P1–P2). The metastasis must have originated from a subclone harboring the stem events only and another subclone with the copy number profile seen in subclone A, indicating polyclonal seeding. The metastasis also has a private copy number neutral imbalance (cnni) of chromosome 4. b NB22 also shows evidence of polyclonal seeding. Samples are from the primary tumor before treatment (B), the primary tumor post treatment (P1–P3), metastases to the lung (M-LU1-2), to the lymph nodes (M-LN1-4), the skull (M-SK), and from the area around the clavicle (M). The stem harbors a 1p cnni, MYCN-amplification and a NF1 deletion. Greek letters denote different structural alterations targeting partly overlapping regions. c WT11 shows subclones present across multiple primary tumor areas (P1–P5). d WT19 displays a similar distribution of subclones as WT11 across the post-treatment biopsies P1–5. e In RMS8, subclones in the biopsies from a local relapse (R) and a distant metastasis (M) form their own branch harboring several additional genetic alterations compared to the primary tumor biopsies (B). The information used to produce the phylogenies can be found in Supplementary Data 1 and 2.