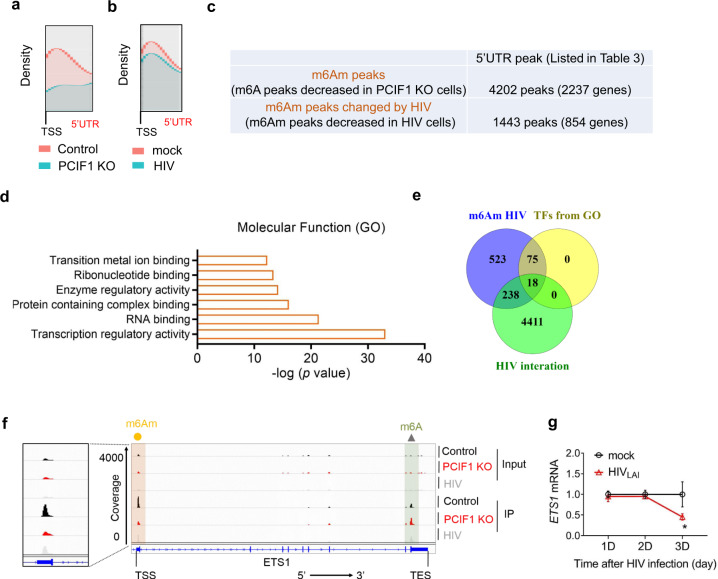
Fig. 3. Identification of m6Am-modified cellular genes altered by HIV infection.
a m6Am-Exo-MeRIP peaks in the 5′UTR are decreased in PCIF1 KO cells. Metagene analysis of m6Am-Exo-MeRIP peaks near the transcription start site (TSS) of all expressed genes in control or PCIF1 KO Jurkat cells. b m6Am-Exo-MeRIP peaks in the 5′UTR are reduced in HIV-infected cells. Metagene analysis of m6Am-Exo-MeRIP peaks in Jurkat cell mock-infected or infected with HIVLAI at an MOI of 4. c m6Am peaks are altered by HIV infection. m6A-MeRIP peaks were aligned to the human genome. And peaks that are located in the 5′UTR and decreased in PCIF1 KO cells were considered as high-confidence m6Am peaks. Of them, peaks that are decreased by HIV infection were shown as potential m6Am peaks changed by HIV. All the peaks from a–c were conserved peaks called from two independent samples. d HIV-changed m6Am genes are enriched in the category of transcriptional factors. Molecular function GO analysis was performed in the 854 m6Am genes changed by HIV using GSEA (http://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp). e Venny plot shown the 18 HIV altered m6Am targeted transcription factors. The 854 HIV-changed m6Am genes were overlapped the 93 transcription regulatory genes and the HIV interaction database (listed in Supplementary Table 4, https://www.ncbi.nlm.nih.gov/genome/viruses/retroviruses/hiv-1/interactions/). f m6Am-Exo-MeRIP peak of ETS1 gene in the 5′UTR is decreased in PCIF1 KO cells and HIV-infected cells. Genome tracks of the ETS1 gene were plotted with called m6A peaks. The m6Am peaks were enlarged in the right panel. One representative of two experiments is shown. g Kinetics of ETS1 mRNA during HIV infection. ETS1 mRNA expression was quantified in MT4 cells infected with HIVLAI at MOI 0.4 for 1, 2, or 3 days. n = 3 independent experiments. Two-sided t-test. Mean ± SD. *p = 0.039.