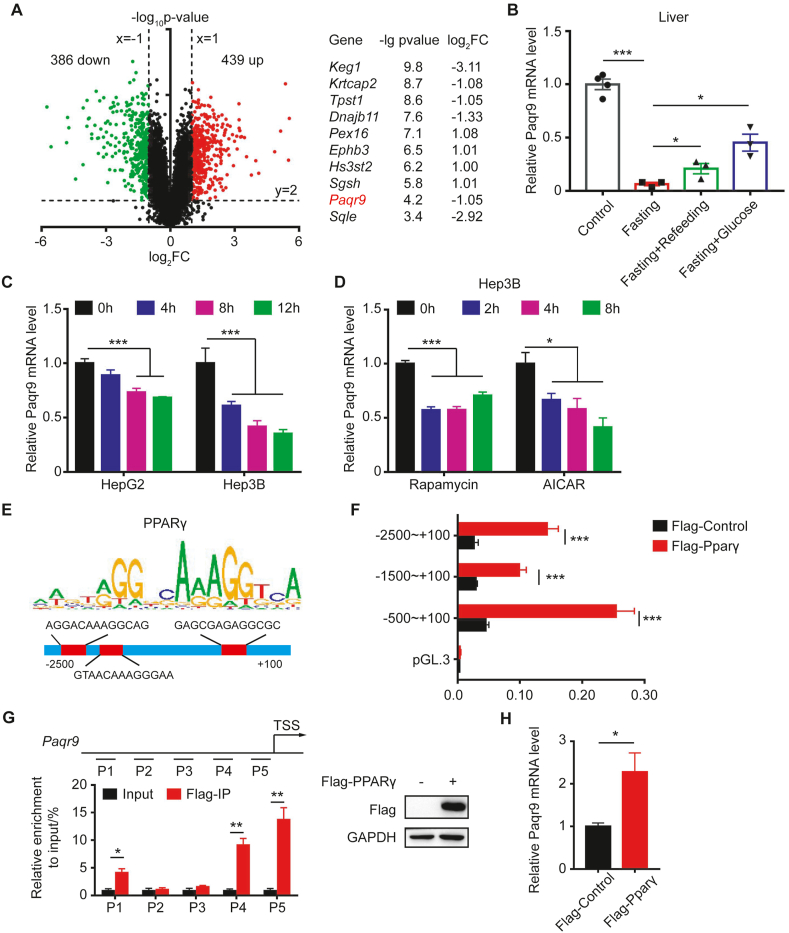
Figure 1.
Paqr9 expression in the liver is regulated by fasting.A. A volcano plot showing RNA-Seq results of liver fasted for 24 hours from GEO dataset GSE92502. Downregulated or upregulated genes were divided by log2–fold change >1 or < −1 with P-value < 0.01. Red dots for upregulated genes and green dots for downregulated genes. The right panel depicts the top 10 most significantly changed genes (except for those involved in lipid metabolism and oxidation pathways). B.Paqr9 mRNA level in the liver after 48 hours fasting and refeeding or peritoneal injection with 10% glucose solution for 3 hours (n = 3 for control and n = 4 for other groups). C.Paqr9 mRNA level in Hep3B and HepG2 cells under starvation condition for different times. D.Paqr9 mRNA level in Hep3B cells treated with 50 nM rapamycin or 1 mM AICAR for different times. E. PPARγ motif analysis and potential binding sites at the Paqr9 promoter. F. Dual-luciferase reporter assay. Different lengths of putative Paqr9 promoter were cloned into luciferase-containing plasmid and transiently expressed in the Hep3B cells along with PPARγ-expressing plasmid as indicated. Relative luciferase activity was calculated by dividing firefly luciferase activity by renilla activity. G. ChIP-qPCR with different regions of Paqr9 promoter. P1 to P5 were five different segments covering the region of −2,500 bp to the transcription start site (TSS) region. Both input and immunoprecipitation results are shown. The expression of Flag-tagged PPARγ is confirmed by Western blotting (right panel). H.Paqr9 mRNA level in Hep3B with overexpression of PPARγ. The mRNA expression results were relative to β-actin in B and relative to α-tubulin in C, D, and H. All the quantitative data are shown as mean ± S.E.M., ∗ for P < 0.05 and ∗∗∗ for P < 0.001.