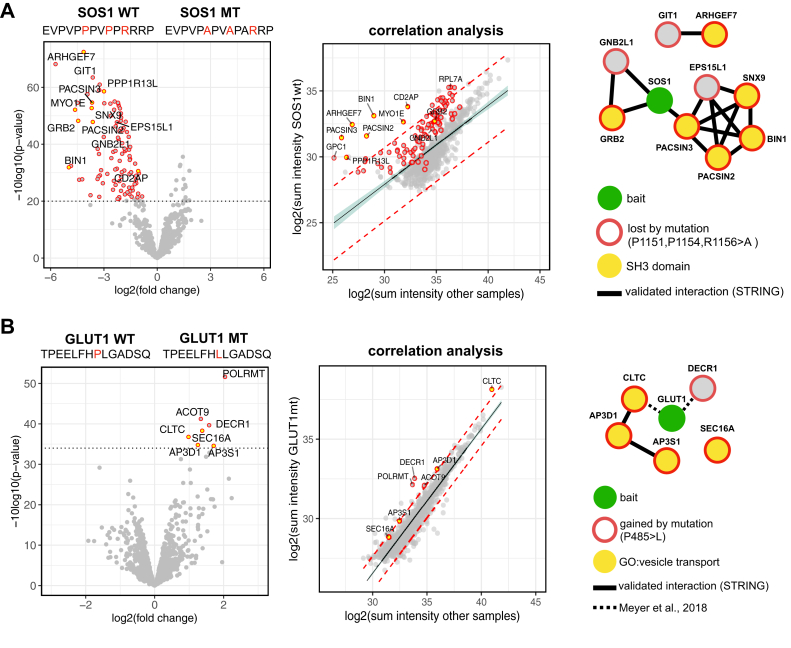
Fig. 2.
PRISMA detects gain and loss of protein binding to mutant peptides.A, mutation of an SOS1 peptide disrupts the SH3-binding motif and causes loss of interactors. B, the disease-causing point mutation in GLUT1 (P485L) creates a dileucine motif that interacts with clathrin and endocytosis adaptor proteins as previously described by Meyer et al., (18). A and B, quadruplicates of PRISMA peptide pull-downs were analyzed with label-free LC-MS/MS. WT and mutated (MT) peptide sequences were compared with a moderated two-sample t test. Volcano plots depict fold changes of detected proteins plotted against their p-value. Significance cut-off (0.05 FDR) is indicated with a dotted line. Correlation plots show log2 (summed intensities) of proteins across peptide spots. Linear regression line (black line) with 95% confidence interval and 95% prediction interval (dotted red line) is indicated. SOS1 interactors detected only in SOS1 WT peptides (SNX9, GIT1, EPS15L1) were removed from the correlation plot in A. Interaction networks display subset of significant interactors connected to the bait in a STRING network or with the GO annotation vesicle transport (A) or SH3 domain (B). Processed data are available in supplemental Table S6. FDR, false discovery rate; PRISMA, protein interaction screen on a peptide matrix.