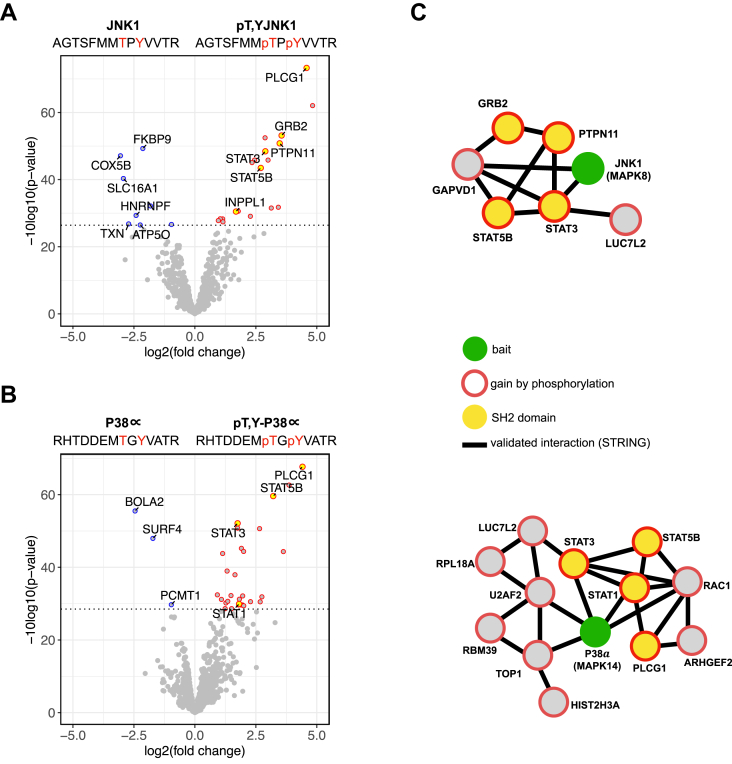
Fig. 3.
PRISMA identifies phosphorylation-dependent interactions of kinase activation loop peptides. Quadruplicates of PRISMA peptide pull-downs were analyzed with label-free LC-MS/MS. Pairwise comparison of unmodified and phosphorylated JNK1 (A) or P38α (B) peptides was performed with a moderated two-sample t test. Volcano plots depict fold changes of detected proteins plotted against their p-value. Significance cut-off (0.05 FDR) is indicated by a dotted line. C, STRING protein interaction networks display interactors gained by phosphorylation and connected to the bait with less than three edges. Processed data are available in supplemental Table S6. FDR, false discovery rate; PRISMA, protein interaction screen on a peptide matrix.