Abstract
The SARS-CoV2 mediated Covid-19 pandemic has impacted humankind at an unprecedented scale. While substantial research efforts have focused towards understanding the mechanisms of viral infection and developing vaccines/ therapeutics, factors affecting the susceptibility to SARS-CoV2 infection and manifestation of Covid-19 remain less explored. Given that the Human Leukocyte Antigen (HLA) system is known to vary among ethnic populations, it is likely to affect the recognition of the virus, and in turn, the susceptibility to Covid-19. To understand this, we used bioinformatic tools to probe all SARS-CoV2 peptides which could elicit T-cell response in humans. We also tried to answer the intriguing question of whether these potential epitopes were equally immunogenic across ethnicities, by studying the distribution of HLA alleles among different populations and their share of cognate epitopes. Results indicate that the immune recognition potential of SARS-CoV2 epitopes tend to vary between different ethnic groups. While the South Asians are likely to recognize higher number of CD8-specific epitopes, Europeans are likely to identify higher number of CD4-specific epitopes. We also hypothesize and provide clues that the newer mutations in SARS-CoV2 are unlikely to alter the T-cell mediated immunogenic responses among the studied ethnic populations. The work presented herein is expected to bolster our understanding of the pandemic, by providing insights into differential immunological response of ethnic populations to the virus as well as by gaging the possible effects of mutations in SARS-CoV2 on efficacy of potential epitope-based vaccines through evaluating ∼40,000 viral genomes.
Keywords: SARS-CoV2, Epitope, MHC alleles, HLA system, Ethnic variations, Immune response
1. Introduction
The ongoing outbreak of severe acute respiratory syndrome (SARS), commonly known as Covid-19, has already infected over 221 million and has led to the death of 4.57 million people worldwide (WHO Coronavirus Disease (COVID-19) Dashboard, n.d.). SARS-CoV2, a newly identified lineage B Betacoronavirus has been held responsible for this global pandemic (Letko et al., 2020). While the current pandemic is unprecedented in scale as compared to the earlier coronavirus outbreaks (Pathan et al., 2020), they are quite alike considering the ongoing quest for an effective preventive/ treatment regime to counter the disease (Ayukekbong et al., 2020). A significant amount of research has been directed at understanding various facets to the pathogenesis. These include the viral attachment and entry (Harvey et al., 2021; Hoffmann et al., 2020; Letko et al., 2020), formation of the coronavirus replication/ transcription complex (Chen, Liu and Guo, 2020; Krichel et al., 2020; Chen, Liu and Guo, 2020, Slanina et al., 2021), replication, transcription and translation of viral proteins (Hillen et al., 2020; Satarker and Nampoothiri, 2020; Wang et al., 2020), virion assembly and release (Kumar et al., 2020; Li et al., 2020) and the commonalities and differences of this disease with seasonal flu and previous SARS infections (Song et al., 2020; Xu et al., 2020). In spite of these efforts, factors affecting susceptibility to infection of SARS-CoV2 and manifestation of Covid-19 are yet to be properly understood and demands more attention. It may be noted that SARS-CoV2’s rate of genomic mutation is similar to most RNA viruses, (De Maio et al., 2021; Mercatelli and Giorgi, 2020; Pathan et al., 2020), and recent evidence seem to suggest that the mortality rates among Covid-19 patients may be associated with the mutation profile of the infecting virus (Toyoshima et al., 2020; “WHO | Variant analysis of SARS-CoV-2 genomes,” n.d.). It has been perceived to be particularly higher for certain SARS-CoV2 variants which have been designated as Variants of Concern (VOC) (Challen et al., 2021; Grint et al., 2021). While a pre-existing medical condition is likely to increase the risk of severity of Covid-19 infection (CDC, 2020), yet another factor influencing the susceptibility to the disease could be the genetic makeup of an individual. The Human Leukocyte Antigens (HLA) system, a major determinant of our ability to detect and neutralize an invading pathogen, is encoded by the Major Histocompatibility Complex (MHC) genes located on chromosome-6. This system has been shown to play a crucial role in the manifestation and outcome of Covid-19 infection (Amoroso et al., 2021, Correale et al., 2020; Langton et al., 2021; Malkova et al., 2021; Pisanti et al., 2020; Shkurnikov et al., 2021). MHCs, which are further categorized into classes-I and II, are highly polymorphic and are known to vary significantly among individuals of different ethnicities. The outcome of an infection event is therefore dependent on both the genotype of the virus as well as the host cell surface (MHC) molecules destined to present viral antigenic peptides to the human T-cell receptor (TCR) of T-lymphocytes (also called killer T-cells or CD8-positive cytotoxic T-cells) and T-helper cells (also called CD4-positive T cells) (Murray and McMichael, 1992, van Montfoort, van der Aa and Woltman, 2014).
In this work, we have investigated how the genetic variations across ethnicities are likely to influence the ability of their immune system in timely recognition of the virus, and in turn, their susceptibility to Covid-19. To this end, state-of-the art bioinformatic tools were used to (a) identify the probable antigenic peptides on the SARS-CoV2 proteomes, and (b) identify HLA alleles which could recognize and present these epitopes to the T-cells, along with the prevalence of these alleles in different ethnic groups. In addition, 40,342 fully sequenced SARS-CoV2 genomes (which were isolated from patients across the globe) were analyzed to probe the possible effect of viral genetic variations on antigenic recognition. Whether the variations in the viral genome over time are likely to change the susceptibility of an ethnic group to Covid-19 infection was evaluated. In this context it may be noted that, given the computational challenges of identifying B-cell epitopes with adequate confidence, the current study did not focus on this aspect of human immunity. Results presented in this work also assumes additional importance when viewed in the context of a recent publication which highlighted the prospect and benefits of considering non-spike proteins for future Covid-19 vaccine designs (Peng et al., 2020).
2. Results
2.1. Identification of SARS-CoV2 epitopes
One of the key mechanisms of identification of an invading pathogen by the host immune system involves MHC class-I and MHC class-II proteins presenting the pathogenic protein fragments (epitopes) on the surface of the CD8-positive cytotoxic T-cells and CD4-positive T-helper cells respectively. Given this, efforts were first made to identify SARS-CoV2 epitopes that could be recognized by the HLA allelic variants. A total of 505 epitopic regions from all the proteins encoded by the SARS-CoV2 reference genome (GenBank Accession no. MN908947) could be identified (see Materials and Methods). Of these, 487 epitopic regions which qualified our criteria for further analysis were predicted to bind to 180 HLA allelic variants (Supplementary Table 1) with reasonable confidence (see Materials and Methods). This comprised of 391 CD8 (MHC class-I) and 96 CD4 (MHC class-II) epitopes. From the list of predicted epitopes and the corresponding HLA alleles, it was observed that some of the HLA alleles could recognize higher number of SARS-CoV2 epitopes and thus might play a more significant role in immune response. Of the 155 MHC class-I associated HLA alleles, that were predicted to be involved in the antigenic recognition of SARS-CoV2 proteins in (CD8-positive) cytotoxic T-lymphocyte cells, the highest number of epitopes were identified by HLA-A*02:11, HLA-B*15:17, HLA-A*24:03, HLA-A*26:02 and HLA-A*68:01 (50, 41, 33, 33 and 26 epitopes respectively). In contrast, among the MHC class-II associated 25 HLA alleles involved in the antigen recognition of SARS-CoV2 proteins in (CD4-positive) T-helper cells, HLA-DRB1×01:01, HLA-DRB1×15:01, HLA-DRB1×15:06 and HLA.DRB1×01:02 were predicted to present the highest number of epitopes (28, 22, 22 and 11 epitopes respectively). It was also noted that few of the epitopes could be recognized by multiple HLA alleles (Supplementary Table 1). It therefore appeared that the potential of a population/ ethnic group to cope with Covid-19 infection could be determined by their MHC gene pool.
2.2. Distribution of SARS-CoV2 cognate HLA alleles across ethnicities
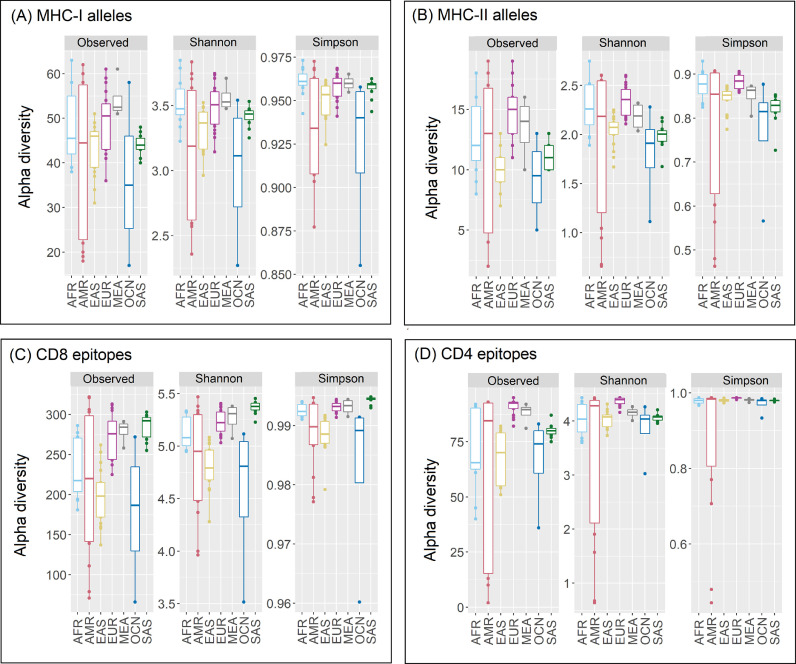
The diversity in the allelic makeup of 82 different ethnic groups constituting seven super-populations were studied using data available from the Allele Frequency Net Database (AFND) and the 1000 genomes project (TGP) (see Materials and Methods and Supplementary Table 2). Only those HLA alleles which occurred with a frequency ≥ 0.01 in at least one of the ethnicities and were predicted to recognize one or more SARS-CoV2 epitopes were considered for the presented analyses. In terms of MHC class-I associated HLA alleles (henceforth termed as MHC-I alleles), Middle East and Africans (MEA) showed the highest richness (Fig. 1 ). Amerindians (AMR) and Oceanians (OCN) had the least MHC-I allelic diversity. In contrast, while Europeans (EUR) and Africans (AFR) demonstrated the highest MHC class-II diversities, South Asians (SAS) and OCN were noted to be least diverse (Fig. 1). The diversity of both MHC-I and MHC-II alleles were found to vary by a large extent among ethnicities comprising the AMR super-population. In general, the HLA allele richness among super-populations was seen to be more diverse in case of MHC-I as compared to MHC-II (Supplementary Table 3). The ethnicities with least MHC-I and MHC-II HLA allelic richness included Melanesian residing in Madang province of Papua New Guinea (OCN_PNG), Mixe and Mixtec Amerindians residing in Mexico (AMR_MXX and AMR_MXY), Amerindians residing near the Gila River in Arizona (AMR_AMX), Aborigine Australians from Cape York Peninsula (OCN_AUS) and Chinese Dai residing in Xishuangbanna (EAS_CDX). In contrast, Hawaiian or other Pacific Islander (OCN_POL), Vietnamese residing in USA (EAS_VUS), Czechs (EUR_CRP), Turks residing in Germany (EUR_GTM) and African Americans (AFR_AFU) exhibited high richness of both classes of MHC alleles. Most others, like the Irish from Southern Ireland (EUR_IRS), English from NW England (EUR_GBN) and Sri Lankans from Colombo (SAS_SLC) demonstrated contrasting characteristics for richness of MHC-I and MHC-II alleles.
Fig. 1.
Diversity of HLA alleles and cognate epitopes across super-populations. Richness (total observed numbers), Shannon diversity and Simpson index indicating the heterogeneity of (A) MHC-I and (B) MHC-II alleles encoded by gene pool of studied super-populations as well as the corresponding (C) CD8-specific and (D) CD4-specific SARS-CoV2 epitopes recognized by the HLA-system in these super-populations. The super-populations are abbreviated as AFR- Africans; AMR- Amerindians; EAS- East Asians; EUR- Europeans; MEA- Middle East and Africans; OCN- Oceanians; SAS- South Asians.
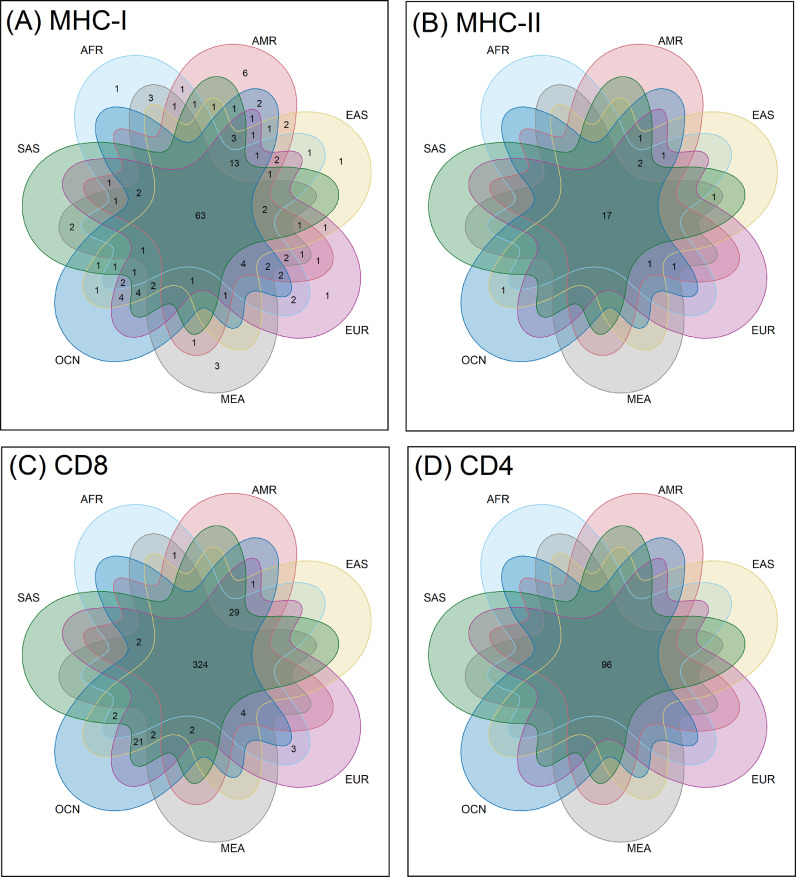
As represented in the Euler diagram of HLA alleles in Fig. 2 (details in Supplementary Table 4), 63 MHC-I and 17 MHC-II alleles were omnipresent in all the super-populations (see Materials and Methods). In addition to this, certain MHC-I alleles specific to each of the super-populations (except SAS and OCN) were also found. MEA lacked 13 MHC-I and 2 MHC-II alleles that were present among all other super-populations. As expected, intra-population variations existed and the occurrence of each of the (SARS-CoV2 associated) MHC-I and MHC-II alleles were not found to be uniform among the samples constituting the ethnic groups (Supplementary Table 3). To account for this, frequency of occurrence for every MHC allele in each of the ethnic groups was computed (see Materials and Methods). Results obtained were used to construct heat maps (Supplementary Figs. 1-2) for gaging the distribution of the potent MHC alleles which could play a role in immune response against SARS-CoV2. While some of the alleles were seen to be more frequent across ethnicities, it was interesting to note that the alleles with the highest antigenic recognition abilities against SARS-CoV2 were often under-represented across different ethnicities. For instance, the most common HLA alleles observed across ethnicities included MHC-I alleles HLA-A*11:01, HLA-A*24:02, HLA-A*02:01, HLA-C*04:01 and MHC-II alleles HLA-DRB1×15:01, DRB1×03:01, HLA-DRB1×07:01. However, amongst them only HLA-DRB1×15:01 (MHC-II) was noted to possess high antigenic recognition ability. In contrast MHC-I alleles (like HLA.A*02:11, HLA.B*15:17, HLA-A*24:03 and HLA.A*26:02) and MHC-II alleles (like HLA-DRB1×01:01 and HLA-DRB1×15:06) with high antigen recognition capability were seen to be less common across ethnicities and in most cases were also sparsely distributed even among samples from same ethnicity. In summary, noticeable variability in the potential of the HLA alleles to recognize and present the SARS-CoV2 epitopes to the immune cells was observed. We subsequently investigated if this could have a bearing on the level of antigenic recognition among different ethnic groups.
Fig. 2.
HLA alleles and cognate epitopes shared across super-populations. Euler diagrams representing the intersecting and unique sets of (A) MHC-I and (B) MHC-II alleles observed in different super-populations as well as the sets of corresponding (C) CD8-specific and (D) CD4-specific SARS-CoV2 epitopes recognized by the HLA-system in these super-populations. The super-populations are abbreviated as AFR- Africans; AMR- Amerindians; EAS- East Asians; EUR- Europeans; MEA- Middle East and Africans; OCN- Oceanians; SAS- South Asians.
2.3. SARS-CoV2 epitope recognition capability of HLA alleles across ethnicities
The overall trend observed with respect to diversity of the predicted CD8 and CD4 epitopes were comparable to those of their corresponding alleles (MHC-I and MHC-II alleles respectively) across ethnicities (Fig. 1). However, certain subtle differences, specifically with respect to their relative richness, were observed. For example, the change in relative richness (number of observed alleles or epitopes for an ethnicity) among South Asian (SAS) to East Asian (EAS) super-populations in the plots associated with MHC-I alleles and the corresponding CD8 epitopes was apparent. Similarly, a change in richness of MHC-II alleles and CD4 epitopes in case of Oceanian (OCN) to EAS super-populations was also visible. The richness of the antigenic recognition potential among Amerindians (AMR) and OCN ethnicities was seen to be quite diverse, particularly for CD8 epitopes (Supplementary Table 3). Most notably, the African Americans (AFR_AFU) had a marked difference in their antigenic recognition potential of MHC-I molecules as compared to the rest of the AFR ethnicities. Similarly, Caribbean Indians (AMR_ACI) showed the highest richness for CD4 epitopes among the AMR super-population. In contrast, Mixe and Mixtec Amerindians residing in Mexico (AMR_MXX and AMR_MXY) and Amerindians residing near the Gila river in Arizona (AMR_AMX) had the least CD8 epitope richness among all the studied ethnicities. Finnish in Finland (EUR_FIN), British from England and Scotland (EUR_GBR) and Utah residents of northern and western European ancestry (EUR_CEU) had a marked difference in CD8 epitopic richness, as compared to the rest of the Europeans. The richness of CD4 epitopes among the East Asian (EAS) ethnicities varied considerably, with Chinese Dai residing in Xishuangbanna (EAS_CDX) having the lowest richness among all the ethnic groups. It was speculated that this change in the relative richness could probably be due to the variations in (a) the distribution of HLA alleles among the ethnic populations, and (b) the disparity in the capacity of some of the HLA alleles to recognize multiple epitopes. To further introspect into this aspect, the richness of MHC-I and MHC-II alleles and their recognition potentials for CD8 and CD4 epitopes among the 82 different ethnic groups were probed in tandem (Supplementary Fig. 3). The mirror-plot (Supplementary Fig. 3) while probing in terms of the ratio of the richness of HLA alleles present (represented along negative x-axis) to the richness of epitopes recognized (positive x-axis) revealed interesting patterns among the various ethnicities. Most EAS and AFR ethnicities, except for Yoruba from Ibadan (AFR_YRI) and Shona community from Harare (AFR_ZMH), had lower to moderate epitope recognition capabilities with respect to the number of MHC-I alleles present. Ethnicities belonging to the SAS super-population exhibited higher ratios of CD8 epitopes identified to MHC-I alleles present. In contrast to the MHC-I alleles, wide variations among the ratio of CD4 epitopes identified to the MHC-II alleles present was observed for the ethnicities comprising each of the super-populations, with AMR sub-populations (ethnicities) being the most diverse.
As indicated in the Fig. 2 (details in Supplementary Table 4), 324 CD8 and 96 CD4 epitopes were found to be common across seven super-populations. While CD4 epitopes were seen to be equally recognized among all super-populations, the CD8 epitopes were found to be differentially recognized. In line with the MHC-I alleles, EAS, EUR and OCN were found to have the highest overlap in terms of recognizing CD8 epitopes. The trends in recognition of epitopes by different alleles present across various ethnicities provided further insights (Supplementary Figs. 4-5). The antigenic peptide FLLPSLATV (Epitope_1 from nsp6) appeared to be the most recognized CD8-specific epitope across ethnicities. Notably, this epitope could be recognized by 15 HLA allelic variants, the highest among all the CD8-specific SARS-CoV2 antigens recognized in this study (Supplementary Fig. 4 and Supplementary Table 1). However, for both the CD8 and CD4 epitopes, there were no observable correlations between the epitopes which were recognized by higher number of HLA variants and those which were most common across ethnicities. For example, while the CD4-dependent antigenic peptides ESPFVMMSAPPAQYE and TQEFRYMNSQGLLPP (Epitope_94 and Epitope_123 respectively) were recognized by five HLA variants each, the maximum among the CD4-specific SARS-CoV2 antigens recognized in this study, (Supplementary Fig. 5 and Supplementary Table 1), Epitope_123 was not as common across different ethnicities as Epitope_94. The above observations indicate that there can be certain discernable differences at an overall population level, with respect to the CD4 and CD8 cell mediated immune response against SARS-CoV2.
2.4. Immune sensitivity to SARS-CoV2 among various ethnic groups
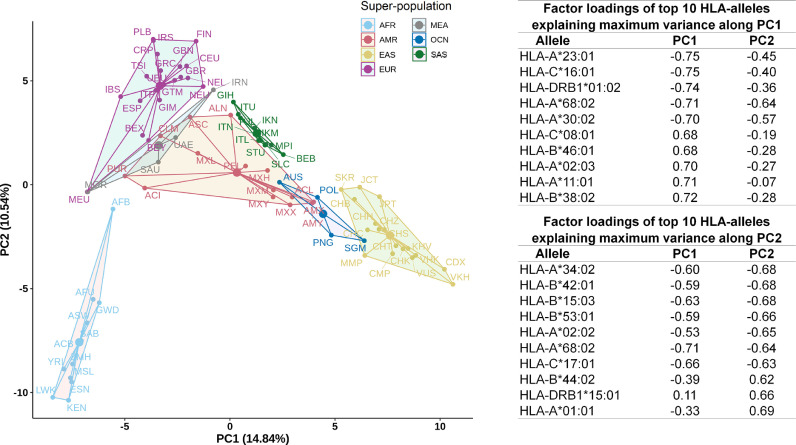
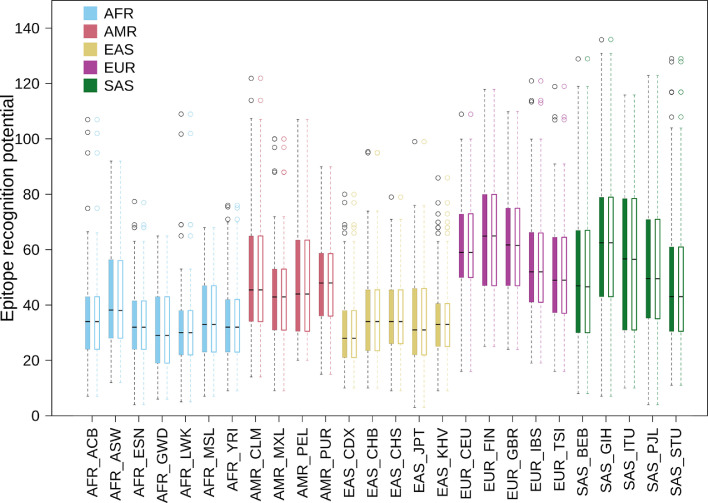
The frequency of occurrence of the 342 HLA alleles (of the classes HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1) constituting AFND and TGP were computed for each of the 82 ethnic groups (Supplementary Table 5). Based on this, the overall epitope recognition potential of the ethnicities and super-populations was computed (see Materials and Methods). Supplementary Figs. 6-7 depict the average count of epitopes recognized by individuals representing super-population and ethnic groups respectively. East Asians (EAS), Africans (AFR) and Oceanians (OCN) reported low potentials to recognize both CD8 and CD4 epitopes. In contrast, ethnicities comprising the European (EUR) and South Asian (SAS) super-populations, showed high potentials to recognize all forms of SARS-CoV2 epitopes. The Peruvians from Lima (PEL) exhibited an interesting pattern with extremely low CD4, but very high CD8 epitope recognition potential. Based on the above observations, we further probed if there were any statistically significant differences between the epitope recognition potential among the various ethnicities and super-populations (see Materials and Methods). The p-value for the one-way ANOVA test among all the super-populations was seen to be less than 2e-16 at 95% confidence interval, thereby implying significant difference in the epitope recognition potential of at least one of the seven super-populations. Indeed, the results obtained from t-test (see Materials and Methods) indicated substantial differences in the recognition potentials of both CD8 and CD4 epitopes among super-populations as well as ethnicities except for AMR and MEA super-population (Supplementary Table 6). Further, to check for any major differences in the epitope recognition potentials between individuals of different ethnicities, a Principal Component Analysis (PCA) was performed (see Materials and Methods). The AFR ethnicities were found to be the most distinct among all super-populations (Fig. 3 ). Further, ethnicities from SAS and EAS formed more compact clusters when compared to ethnicities from AMR and EUR, with ethnicities from AMR being the most dispersed. The epitope recognition potentials of the Mexican and the USA appeared to be distinct from the rest of the Amerindians. Aborigine Australians (AUS) and the Malay from Singapore (SGM) were found to cluster more closely with ethnicities of other super-populations. AUS and SGM clustered with ethnicities of AMR and EAS super-population respectively. While the prevalence of specific HLA alleles in the gene pool of a population or an ethnic group provides an overall idea about how well recognized the SARS-CoV2 epitopes would be in that population, the immune response of each of the individuals would be independently governed by their own allelic make-up. The above results provide an average estimate of (CD4 and CD8 cell mediated) immune sensitivity to SARS-CoV2 for individuals belonging to a population and highlights inter-ethnic differences therein.
Fig. 3.
Principal component analysis (PCA) of epitope recognition potential. PCA based on frequency distribution of HLA alleles in different populations. Only those HLA alleles which could recognize the predicted SARS-CoV2 epitopes were considered. The variance explained by each of the first two principal components (PC1 and PC2) are indicated in brackets beside respective axis labels. Factor loadings of key HLA alleles explaining maximum variance along the first two principal components are also tabulated.
2.5. Impact of genetic variability of SARS-CoV2 on immune recognition
In addition to the HLA makeup, yet another factor which could determine the fate of the infection process is the genetic variation in the SARS-CoV2 genomes. Genetic variation could lead to epitope modifications, which in turn, might alter the binding affinity/ recognition of the viral epitopes by MHC-I/II alleles. To accommodate for this factor, the epitope signature of 40,342 SARS-CoV2 genomes, were generated (see Materials and Methods) based on the 487 epitopes predicted for the reference genome and their 4194 variants, henceforth referred to as variants of ‘reference’ epitopes (VREs) (Table 1 and Supplementary Table 7). Apart from the original 487 epitopes, only 25 of the 4194 VREs were seen to be present in more than 0.5% of the studied SARS-CoV2 genomes (more frequent VREs or MFVREs). Moreover, some of the variants were found to exclusively co-occur among a sub-set of strains isolated from certain geographies, most prominently among those isolated from India or United Kingdom. Even more significantly, a particular variant (SEVGPEHSL at position 376 on ORF1a) was noted to be exclusively absent among the SARS-CoV2 genome sequences isolated and sequenced in Iran (Supplementary Table 7). It was further observed that while the MFVREs (those observed in more than 0.5% of the studied SARS-CoV2 genomes) exhibited a mixed pattern of altered immunological behaviors, the human MHC-I/II alleles have a lower binding affinity/ recognition potential for a large proportion (2072 out of 4169) of the lesser frequent VREs (LFVREs) (Table 1).
Table 1.
Overview of variations observed in predicted epitopic regions across different SARS-CoV2 genomes .
| Number of genomes analyzed | 40,342 |
| Number of epitopes (RE) predicted from the reference genome (NCBI Genbank accession no: MN908947) | 487 |
| Number of variants of ‘reference’ epitopes (VRE) identified among the analyzed genomes | 4194 |
| VREs present in at-least 0.5% of the studied genomes more frequent VREs (MFVRE) | 25 |
| VREs present in less than 0.5% of the studied genomes less frequent VREs (LFVRE) | 4169 |
| VREs recognized by HLA system (VE) | 2610 |
| VEs with no change in epitope recognition by HLA alleles w.r.t. REs (6 MFVREs + 1588 LFVREs) |
1594 |
| VEs recognized by higher number of HLA alleles w.r.t. REs (7 MFVREs + 509 LFVREs) |
516 |
| VEs recognized by fewer number of HLA alleles w.r.t. REs (3 MFVREs + 497 LFVREs) |
500 |
| VREs not recognized by HLA system (VX) (9 MFVREs + 1575 LFVREs) |
1584 |
| Total number of epitopes across all genomes (RE + VE) | 3097 |
While it would be interesting to understand the effect of these variations in the viral genome on the antigenic recognition potentials among different ethnicities, no major effects were expected, given that only 25 of the variants (MFVREs) were present in at least 0.5% of the studied SARS-CoV2 genomes. Even in the hypothetical scenario, wherein every population/ ethnicity is simultaneously exposed to all the 40,342 viral variants, the individual antigenic recognition potential would be driven by the originally identified 487 epitopes, and not by the LFVREs. Supplementary Table 8 lists the HLA alleles which could recognize the 25 MFVREs or their corresponding reference epitopes. The most frequently observed MFVREs were STVFPLTSF and TVFPLTSFGPLVR (variants of Epitope_461 and Epitope_479 respectively), both from the NSP12 coding region of the SARS-CoV2 genome and are present in over 73% of the studied genomes. In case of STVFPLTSF, the allele HLA-B*15:17 was found to recognize both the reference as well as the variant epitope. In addition to this, HLA-A*26:02 could recognize the variant epitope, but not the reference epitope. HLA-A*26:02 has the maximum observed frequency among the Japanese in Tokyo (EAS_JPT) (allele frequency 2.4%), but at the same time not observed in 74 of the 82 sub-populations studied (Supplementary Table 8). Therefore, the recognition of the variant epitope by an additional HLA allele is expected to have a limited impact, if any, in a small population group. On the other hand, in case of the variant TVFPLTSFGPLVR, while the HLA-A*68:01 allele could recognize the reference epitope, the variant was no longer recognized. While HLA-A*68:01 is observed in maximum frequency in the AMR_MXM ethnicity (14.5%) and other closely related Amerindian populations, it was noted to be present in all but 8 of the 82 sub-populations considered (Supplementary Table 8). So, one may expect that the variant epitope escaping recognition by HLA-A*68:01 could be a contributor in altered immune response. However, HLA-A*68:01 has earlier been reported as one of the strongest binders of most viruses (Barquera et al., 2020) and it is likely that the loss of recognition of a single potential epitope may not influence its overall binding affinity to the virus.
Considering the continuously evolving genome of SARS-CoV2, which entails selection/ retention of specific epitope variants over time in the newly emerging genomes, an analysis was performed to check if there were any temporal changes in the susceptibility to Covid-19 in any of the ethnicities (see Materials and Methods). Even in this case (Fig. 4 ), it was noted that temporal variations in the SARS-CoV2 genome, while resulting in certain changes in its epitope signature, did not appreciably alter the epitope recognition ability among ethnic groups, at least at a population level (also see Supplementary Fig. 8). Since ethnic groups (and people from same geography) are known to predominantly encode for certain HLA gene variations, it may be perceived that the SARS-CoV2 epitope recognition potential of an ethnic group is not likely to change owing to the temporal variations in the viral genome. Further, it will be appreciated that although the said changes in the viral genomes might alter the biological functioning of the viral genes, yet they remain largely inconsequential with respect to immune determination in the host.
Fig. 4.
Average epitope recognition potential against evolving SARS-CoV2 variants. Representation of the average of predicted SARS-CoV2 epitope recognition potential of individuals from different populations (ethnic groups), considering different SARS-CoV2 genome variants observed across geographies. For each population (ethnic-group), there are two box-plots indicating their average ‘weighted’ epitope recognition potential (see Materials and Methods). For each population, the color-filled box on the left corresponds to the epitopes predicted from the first 10,000 SARS-CoV2 genomes (out of a total 40,342 genomes analyzed in the study) when arranged in the chronological order of their collection dates. The corresponding unfilled boxes on the right corresponds to the epitopes predicted from the last 10,000 SARS-CoV2 genomes based on their collection dates. Although the SARS-CoV2 genomes may be evolving over time, no observable variation in the way its epitope repertoire is recognized by individuals at an overall population level could be noted. Each data-point on the plot represents the average of the number of epitopes in a given SARS-CoV2 genome that could be identified by all the individuals in an ethnic population. Dark black lines, for each of the ethnic groups, indicate the mean value for these data-points for the studied 40,342 SARS-CoV2 genomes. HLA data from 1000 genomes project (TGP) database, which provided individual specific allelic information, was used in this analysis. A total of 505 predicted epitopes from the reference SARS-CoV2 genome along with 2716 epitope variants identified across 40,342 genomes recognized by 292 HLA alleles were used to calculate the individual epitope recognition potential. Super-population names are abbreviated as AFR- Africans; AMR- Amerindians; EAS- East Asians; EUR- Europeans; SAS- South Asians, and prefixed to each of the population names in the plots, as well as represented with boxes drawn in specific colors. The abbreviations used for names of different populations/ ethnic groups are provided in Supplementary Table 2.
3. Discussions
There have been three major coronavirus associated SARS outbreaks in the last 20 years. While there have been extensive research regarding the pathophysiology of these viruses (Chen, Liu and Guo, 2020; Chen, Liu and Guo, 2020, Harvey et al., 2021; Hillen et al., 2020; Hoffmann et al., 2020; Krichel et al., 2020; Kumar et al., 2020; Letko et al., 2020; Li et al., 2020; Satarker and Nampoothiri, 2020; Slanina et al., 2021; Song et al., 2020; Wang et al., 2020; Xu et al., 2020), as yet we have limited knowledge into the factors affecting susceptibility to SARS-CoV2 infection and manifestation of Covid-19. Some scientists have opined that the mortality rate in Covid-19 could be linked to the genomic profile of the infecting virus (Challen et al., 2021; Grint et al., 2021; Toyoshima et al., 2020; “WHO | Variant analysis of SARS-CoV-2 genomes,” n.d.) which seem to mutate at a rate similar to most RNA viruses (De Maio et al., 2021; Mercatelli and Giorgi, 2020; Pathan et al., 2020). Further, an underlying/ pre-existing medical condition might be considered to be an added risk towards increasing the severity/ complications associated with Covid-19 infection (CDC, 2020). In addition, the host genetic makeup could play a decisive role in disease manifestation. In this context, polymorphic genes like the Human Leukocyte Antigen (HLA) system which is known to vary significantly among individuals of different ethnicities assumes importance. Thus, the outcome of an infection event is multi-factorial and at least dependent on (a) genotype of the virus and (b) host cell surface (MHC) molecules destined to present viral antigenic peptides to the human immune cells.
To understand this, the antigenic peptides from over 40,000 SARS-CoV2 genomes were predicted for all major HLA alleles (viz. HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1), as documented in the Allele Frequency Net Database (AFND) and the 1000 genomes project (TGP). The state-of-the-art tools NetMHCpan 4.1 and NetMHCIIpan 4.0 were used for the purpose in the current work (Reynisson et al., 2020). It may be noted that most of the other available tools and web-services for predicting MHC-I and MHC-II epitopes (Jespersen et al., 2017; Paul et al., 2016, Zhang, Lund and Nielsen, 2009, Saha et al., 2004) were not equipped to handle the entire HLA allelic set that had been reported in AFND and TGP. Further, most of the other tools also did not have a provision to identify variable length MHC-I epitopes. Consequently, some of the earlier studies associated with in silico identification of SARS-CoV2 epitopes (Abdelmageed et al., 2020; Dong et al., 2020; Lin et al., 2020; Naz et al., 2020; TOPUZOĞULLARI et al., 2020) are likely to have suffered from the mentioned limitations associated with these tools (Jespersen et al., 2017; Paul et al., 2016, Zhang, Lund and Nielsen, 2009, Saha et al., 2004). In other words, these studies might not have arrived at a list of epitopes as comprehensive as the one mentioned in the current study. Further, studies following a combinatorial approach (H.-Z. Chen et al., 2020; Zaheer et al., 2020) are also expected to miss some of the epitopes reported in this study. It may however be noted that like most previous studies, the current study refrained from predicting potential B-cell epitopes, due to the computational complexities involved in the process and may be considered as a limitation of this exercise. Most B-cell epitopes are conformational in nature and comprise of discontinuous amino acid stretches which are difficult to be identified (with adequate confidence) from genomic information alone (Wang et al., 2011).
A total of 391 CD8 (involving 155 MHC class-I alleles) and 96 CD4 (involving 25 MHC class-II alleles) SARS-CoV2 epitopes were predicted in the current study. Given the in silico nature of the work, the immunogenicity of the predicted epitopes could not be verified experimentally. However, it may be noted that the prediction algorithms were trained on experimentally validated epitopes and therefore expected to reflect the immunological interplay involving the human MHC alleles and the viral epitopes. The highest number of SARS-CoV2 epitopes were presented by the MHC class-I associated HLA alleles HLA-A*02:11, HLA-A*24:03, HLA-B*15:17, HLA-A*26:02 and HLA-A*68:01, and MHC class-II associated HLA allelesHLA-DRB1×01:01, HLA-DRB1×15:01, HLA-DRB1×15:06, HLA-DRB1×01:02. The diversity of both HLA alleles encoded in the genome as well as the epitopes recognized by their HLA systems was observed to vary across the super-populations (Fig. 1) and among the ethnic groups (Supplementary Table 3) comprising these super-populations. While the super-populations were largely seen to cluster in terms of the MHC-I genes encoded in the genome, such patterns could not be observed for MHC-II genes (Supplementary Figs. 1–2). In addition, certain HLA alleles were found to be more efficient in recognizing SARS-CoV2 epitopes as compared to others. Given the skewed distribution of these HLA alleles among a few of the ethnic groups, contrasting characteristics with respect to the ratio of the number of epitopes to the number of HLA alleles involved in their recognition, among each of the ethnic groups were observed, even among ethnicities from the same super-population (Supplementary Fig. 3).
The role played by HLA genes (differentially occurring among ethnicities/ super-populations) in the SARS-CoV2 immune response is under-explored with respect to other aspects affecting the manifestation and outcome of the Covid-19 infection. Some of the available reports are mentioned herein with respect to our presented results. Our results concur with the report on the susceptibility of genotypes to predisposition of coronavirus infection (Malkova et al., 2021). The genotypes with lower susceptibility (viz., HLA-A*02:02, B*15:03, C*12:0) were seen to recognize higher number of epitopes than the high susceptibility genotypes (viz., HLA-A*25:01, B*46:01, C*01:02). The HLA-C*04:01 allele has been reported to be associated with an increased risk of intubation and severe clinical symptoms of Covid-19 (Increased risk of severe clinical course of COVID-19 in carriers et al., 2021). We found that this could recognize only two SARS-CoV2 epitopes, thereby hinting at a potential vulnerability to Covid-19 infection among individuals harboring this gene. HLA-C*04:01 was found to be most frequent among African (AFR), followed by Amerindian (AMR) and Oceanian (OCN) super-populations. We were intrigued by the contradicting literature evidence on the relationship of HLA-A*02:01 to the risk of infection (Migliorini et al., 2021; Shkurnikov et al., 2021). Given its widespread occurrence (one of the most frequent HLA genes in most of the super-populations), we believe additional research is required for ascertaining its relation to the disease outcome. In a study conducted in Italy, HLA-C*06:02 was shown to be better correlated with the patient group, indicating at its association with disease severity (Novelli et al., 2020). In this regard, the higher abundance of HLA-C*06:02 among the Middle East and African (MEA) and South Asian (SAS) super-populations assume importance. Among the MHC-II alleles HLA-DRB1×03:01 was observed to be the second most frequent allele in AMR and MEA. This allele has been earlier reported to increase the risk of infection (Shen et al., 2021). HLA-DRB1×01:01 and HLA-DRB1×15:01 was observed to be able to recognize highest number of CD4 epitopes in all seven super-populations. Noticeably, HLA-DRB1×01:01 has been shown to be negatively associated with mortality rate of hospitalized patients (Romero-López et al., 2021) and was found to be most abundant among European (EUR) super-population. Yet another study reported a significantly lower frequency of the haplotype DQA1×01:01‐DQB1×05:01‐DRB1×01:01 among the asymptomatic group when compared to patients with high severity (Langton et al., 2021). In contrast to our observations, this would indicate at a diminished capability to identify and counter SARS-CoV2 invasion among individuals harboring HLA-DRB1×01:01. Given that the haplotype information was not available in AFND, the above could not be pursued further. For the same reason, we could not compare our results with some of the other literatures which had used HLA haplotype data for drawing inference (Khor et al., 2021; Pisanti et al., 2020).
In this study, an attempt was further made to gage the variations in the SARS-CoV2 epitope regions resulting from temporal changes in the viral genome and its effect on the epitope recognition potential across the ethnicities worldwide. The SARS-CoV2 genomic variation data which was obtained for different geographies over a six-month period indicated that although the SARS-CoV2 genomic variations altered the overall predicted SARS-COV2 epitope signature profile (Table 1 and Supplementary Table 7), most of the alternate epitopes were not as common and occurred in ≤ 0.5% of the analyzed genomes. Further, a comparative study of the variations observed in the SARS-CoV2 epitopes when compared to the variations observed spanning the protein lengths did not indicate any selective pressure which may be at play to evade the immune response (see Supplementary File 1 and Supplementary Fig. 9). The overall capacity of an ethnic group to recognize SARS-CoV2 did not seem to change either (a) on encountering a new viral strain (Fig. 4) or (b) over the course of the pandemic (Supplementary Fig. 8). On another note, when we analyzed the possible effects of the characteristic mutations defining the SARS-CoV2 Variants of Concern (VOC) as obtained from the PANGO lineages server [hosted at cov-lineages.org] (O'Toole et al., 2021; Rambaut et al., 2020) on the predicted reference epitopes, we observed a reduction in the number of HLA alleles which could recognize variants of the epitopes when compared to the allelic repertoire recognizing the reference epitopes (Supplementary Table 9). For instance, in case of the Alpha (B.1.1.7) variant, out of three epitope variants, two showed a decrease in the number of alleles that could recognize the epitopes, whereas one of the variants showed an increase in the number of alleles that could recognize it. Four out of five variants in Beta (B.1.351) and three out of four variants in Gamma (P.1) were also observed to be recognized by lesser number of alleles. As for Delta (B.1.617.2) strain, none of the four variants of the reference epitope could be recognized by the HLA alleles. It may appear that reduction in the number of HLA allele capable of identifying the mentioned epitope variants in the VOCs would aid the virus to evade the host immune system. However, it would be difficult to comment on the aspect given that the set of mutations defining a VOC are only affecting a handful of predicted epitopes (a total of 15 for the four VOCs taken together) with respect to the total set of 487 predicted epitopes in the reference genome. Elaborate genetic and epidemiological studies would be required to derive any conclusive evidence on the above aspects. Nonetheless, our study provides initial clues and intriguing insights relevant to host-virus interactions.
Moreover, a minimal set of SARS-CoV2 epitopes was identified, which can be recognized by the HLA repertoire of individuals from all ethnicities (Table 2 ). Candidates from this set of predicted antigenic peptides could provide an opportunity to design newer vaccines against Covid-19 (refer to Supplementary File 1 and Supplementary Table 10). While most of the currently available Covid-19 vaccines are designed to target the viral spike protein, a recent study while inspecting T-cell memory from patients recovering from Covid-19 infection commented on the prospect of considering non-spike proteins within future Covid-19 vaccine designs (Peng et al., 2020). Moreover, the outcome of this study can also aid in disease prognosis and designing of personalized therapy regimes for Covid-19 patients. The idea of providing personalized therapy to Covid-19 patients, especially those requiring critical care is already under clinical consideration (Cacciapuoti et al., 2020, Fang and Schooley, 2021, Garcia-Vidal, 2020). In addition, results presented herein could also prove beneficial in understanding/ countering a future flu/ pneumonia outbreak involving a similar lineage B Betacoronavirus. Given that the world has already witnessed two such major outbreaks in less than 20 years, it would be prudent to prepare blueprints of a more potent coronavirus vaccine, particularly against the lineage B Betacoronavirus, before the next outbreak.
Table 2.
List of SARS-CoV2 antigenic peptides relevant for potential multi-epitope vaccine design. The table comprises of 11 CD8-specific and 17 CD4-specific epitopes that may be considered as potential multi-epitope vaccine candidates based on their recognition by the HLA repertoire prevalent among all the seven super-populations. The position of these peptides on the respective SARS-CoV2 proteins is also mentioned. The list is sorted based on descending order of VaxiJen (Doytchinova and Flower, 2007) scores. The presence of these peptides in SARS coronavirus proteome is also indicated.
| Epitope Sequence | Position | VaxiJen Score | Present in SARS |
| CD8-specific | |||
| VYVLQSINF | ORF3a-112–120 | 1.247 | – |
| VYFLQSINF | ORF3a-112–120 | 1.1117 | – |
| NYMPYFFTL | NSP3–1349–1357 | 1.0015 | – |
| SLSKGVHFV | ORF3a-72–80 | 0.9828 | – |
| IYNDKVVGF | NSP12–37–45 | 0.9104 | – |
| ALFKGVHFV | ORF3a-72–80 | 0.9039 | – |
| FLLPSLATV | NSP6–70–78 | 0.5954 | Yes |
| ALSNGVHFV | ORF3a-72–80 | 0.5551 | – |
| FLLPSFATV | NSP6–70–78 | 0.4627 | – |
| YYTSNPTTF | NSP3–718–726 | 0.1415 | – |
| IYNDKVAVF | NSP12–37–45 | 0.1096 | – |
| CD4-specific | |||
| GTWLTYTGAIKLNDK | Nucleocapsid-328–342 | 1.1129 | – |
| GTWLTYTGAIKLDDK | Nucleocapsid-328–342 | 0.9934 | – |
| FTGYRVTKNSKIQIG | NSP13–182–196 | 0.8361 | – |
| FTGYRVTKNSKVQIG | NSP13–182–196 | 0.7725 | Yes |
| SGTWLTYTGAIKLND | Nucleocapsid-327–341 | 0.7572 | – |
| AVVYRGTTTYKLNVG | NSP13–208–222 | 0.7324 | Yes |
| SGTWLTYTGAIKLDD | Nucleocapsid-327–341 | 0.6215 | – |
| PSDFVRATATIPIQA | ORF3a-25–39 | 0.5466 | – |
| SDFVRATATIPIQAS | ORF3a-26–40 | 0.5411 | – |
| LGTWLTYTGAIKLDD | Nucleocapsid-327–341 | 0.5123 | – |
| CDAVVYRGTTTYKLN | NSP13–206–220 | 0.4598 | Yes |
| GDAVVYRGTTTYKLN | NSP13–206–220 | 0.4083 | Yes |
| DAVVYRGTTTYKLNV | NSP13–207–221 | 0.3576 | Yes |
| PSDFVRATATIPIPA | ORF3a-25–39 | 0.2256 | – |
| SDFVRATATIPIPAS | ORF3a-26–40 | 0.1634 | – |
| NKHIDAYKTFPSTEP | Nucleocapsid-354–368 | 0.0081 | Yes |
| DGSIIQFPNIYLEGS | NSP4–195–209 | −0.0387 | – |
The context and results of the presented work is somewhat aligned to another intriguing aspect of the prevailing pandemic with respect to disease severity and case fatality rate (CFR). Appreciable spatial and temporal variations in CFR have been observed across geographies and suitable explanation(s) for these variations have eluded researchers, given the wide array of possible confounding factors. A key observation in this regard pertains to the economic status of a country. Death rates due to Covid-19 has been observed to be positively associated with GDP (per-capita) in multiple studies (Cao et al., 2020; Sorci et al., 2020). Higher death rates despite (expected) better access to healthcare in high-income populations seems outright counter intuitive. While some scientists have tried to explain these observations citing higher life-expectancy and consequently an older population in high-income countries, who would be more susceptible to Covid-19, others have hinted at the possibility of the "so called hygiene-hypothesis" at play (Bloomfield et al., 2016, Chatterjee, Karandikar and Mande, 2021). The results of the current study unravel yet another interesting aspect in this context pertaining to the genetic diversity. Severe cases of Covid-19 disease have been seen to be characterized by higher levels of inflammatory cytokines and CD8+ T cell exhaustion (Yang et al., 2020). Reports have also indicated that in case of milder Covid-19 infections, not leading to death and other complications, higher proportions of SARS-CoV2 specific CD8+ T cells have been identified (Peng et al., 2020). Our results indicate that the heavily affected European population (EUR) tends to harbor a larger fraction of MHC-II alleles in their gene-pool, which are specific to SARS-CoV2 epitopes. On the other hand, the South Asian (SAS) population, having a relatively lower fatality rate, exhibited a relatively larger proportion of MHC-I alleles specific to the SARS-CoV2 epitopes. It is worth further investigation whether a larger pool of MHC-I alleles presenting SARS-CoV2 antigens to CD8+ T cells in the SAS population indeed can be linked to the apparently lower fatality rate in this region. A recent study while reporting an inverse association between MHC-I epitopes and mortality rates also indicated at this possibility (Wilson et al., 2021). Moreover, any possible relationship of the larger pool of the MHC-II alleles in the EUR population with CD4+ T Cell activation, cytokine production leading to a disproportionate immune response (or a so called "cytokine-storm") will also be intriguing to explore. While the current work is limited by the number of representative individuals genotyped for each population in AFND and TGP, the results nonetheless provide an overview of possible trends in protective immunity and T-cell responses against SARS-CoV2 across different geographies/ ethnicities.
Overall, to our knowledge, this is the first ever account to capture the effects of the evolution of SARS-CoV2 genome on its potential interactions with the human HLA gene products in a global perspective. This assumes immense importance in the context of vaccine development and its efficacy against the newer lineages of SARS-CoV2. It highlights the need of understanding the crosstalk of the pathogen with the components of HLA system could have far reaching consequences in our coexistence with SARS-CoV2 and other RNA viruses which we may encounter in future. It is pertinent to note that the nature of our findings, obtained through a bioinformatic/computational exercise, were dependent on (a) the availability of data in public repositories and (b) the efficiency of the available bioinformatic tools. As a result, insights into some of the aspects of immune response, such as, if there are any B-cell specific conformational epitopes in SARS-CoV2, expression levels of the MHC alleles in response to Covid-19 infection and its variations across ethnicities, age groups, co-morbidities, etc., if any, could not be obtained. Furthermore, while it would have been ideal to validate the derived conclusions in an experimental setting, such an exercise lies beyond the scope and reaches of the current endeavor given the scale of the project, which involved analyses of over 40,000 viral genomes and the HLA gene data of nearly 150,000 individuals from 82 ethnicities spread across the globe. Nonetheless, the perspectives presented are interesting and quite pertinent with respect to the ongoing pandemic and deserve to be shared with the larger scientific community. We hope that a larger collaboration/ consortium may be forged in the days to come towards validating the insights derived in silico.
4. Materials and methods
4.1. Data acquisition
Full length sequences of SARS-CoV2 proteins (reference sequences) were obtained from NCBI (GenBank Accession no. MN908947). Further, individual non-structural protein sequences were also obtained from NCBI (NCBI Accession no. YP_009725300.1 to YP_009725312.1). These protein sequences were used for predicting antigenic peptides (epitopes) on the viral proteome. In addition, high quality fully sequenced genome sequences of SARS-CoV2 were obtained from GISAID (https://www.gisaid.org/) (Elbe and Buckland-Merrett, 2017, Shu and McCauley, 2017). Genomic data and corresponding metadata for 40,342 genomes (which were deposited to GISAID till 11th June 2020) have been used in this analysis (see Supplementary File 2 for details).
The allele frequency distributions for different ethnicities were obtained from AFND - Allele Frequency Net Database (http://www.allelefrequencies.net/). The ‘Gold standard’ allele frequency data belonging to the 12 geographical regions were obtained and filtered to remove (a) populations wherein the ethnic origins were marked as ‘Mixed’ or were not clearly mentioned and (b) wherein the information pertaining to the one of the five major HLA classes (as suggested by AFND, viz. HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1) (Gonzalez-Galarza et al., 2020) were missing. Finally, allele frequency data corresponding to the major MHC-I and MHC-II HLA types were retained for 56 ethnicities. Furthermore, the human HLA allelic profiles were also obtained from TGP - 1000 genomes project (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/HLA_types/). This data corresponded to 2693 individuals (Samples) who belonged to five super-populations (Regions) and 26 ethnicities (Populations) as described in the 1000 genomes project (Abi-Rached et al., 2018). Data belonging to 13 samples of the TGP which had 'None' mentioned in one or more of the allele columns, were not considered for the analysis. For TGP, ‘frequency’ of an allele in each ethnicity was computed as number of individuals (Samples) from that ethnicity which encoded for the allele divided by the total number of individuals (Samples) representing the ethnic group. The combined set of MHC-I and MHC-II alleles from AFND and TGP, were filtered to remove all alleles which had an abundance < 0.01 in all the studied ethnicities. The final allele set comprised of 342 alleles (Supplementary Table 5) belonging to 82 ethnic groups that were arranged into seven super-populations (Supplementary Table 2).
4.2. Epitope prediction
NetMHCpan 4.1a (http://www.cbs.dtu.dk/services/NetMHCpan/index_4.1a.php) and NetMHCIIpan 4.0 (http://www.cbs.dtu.dk/services/NetMHCIIpan/) were used with default parameters for identifying strong and weak binders (i.e., 0.5 and 2 for NetMHCpan and 1 and 5 for NetMHCIIpan) (Reynisson et al., 2020) to generate epitopes from SARS-CoV2 protein sequences against 258 MHC-I and 84 MHC-II alleles obtained from AFND and TGP. Variable length (8 to 14-mer) CD8 epitopes (mapping to MHC-I alleles) and 15-mer CD4 epitopes (corresponding to MHC-II alleles) were thus obtained which were further filtered for subsequent use. First, all predictions with scores < 0.95 were discarded to retain only epitope HLA allele pairs with high binding affinity (recognition capacity). Next, in case of a given MHC-I allele, if one CD8 epitope was found to be a sub-strings of another, then the predicted CD8 epitope with the highest prediction score was retained. This final list comprised of 487 (391 CD8 and 96 CD4) epitopes having high prediction scores for 180 (155 MHC-I and 25 MHC-II) HLA alleles was used for further analysis (Supplementary Table 1). In addition, the epitopes corresponding to the less abundant MHC-I and MHC-II alleles in the TGP were also predicted using the above protocol. This data was used for a subset of analysis (see Fig. 4).
4.3. Computing allele and epitope distribution across populations
All the analysis, computations and visualizations were performed using R version 4.0. The allele frequency distributions across ethnicities (Supplementary Table 5) and the epitope HLA allele pairs (Supplementary Table 1) were used to compute various diversity indices (viz. richness, Shannon index and Simpson index) for MHC-I and MHC-II alleles as well as CD8 and CD4 epitopes. The tidyverse package version 1.3.1 was used for all the data wrangling operations. While the phyloseq package version 1.32.0 was used to calculate diversity, the ggplot2 package version 3.3.3 was used for the visualizations. The data was also used to generate Euler plots to depict the distribution of alleles across super-populations as well as the epitopes which would be recognized by these alleles. For this purpose, the venn package version 1.10 was used. Moreover, heatmaps to visually inspect the frequency of distribution of HLA alleles (both MHC-I and MHC-II) and the identified epitopes across the 82 ethnic groups were created using the heatmap.2 function of gplots library - version 3.0.3 (Supplementary Figs. 1–2, 4–5). For this purpose, recognition potential of an identified epitope for a given ethnicity was computed as the cumulation of the frequencies of all alleles observed in that ethnicity which could recognize the epitope. The heatmaps were further augmented with a color-key along the vertical axis depicting (a) the number of epitopes recognized by the HLA alleles (Supplementary Figs. 1-2), and (b) the number of HLA alleles which could identify a given epitope (Supplementary Figs. 4-5) according to the data provided in Supplementary Table 3. In addition, the data represented in the heat maps were also used to hierarchically cluster the ethnic groups based on the HLA alleles they encode and the epitopes they could identify (Supplementary Figs. 1–2, 4–5). The hclust function from stats package was used for this purpose. The average epitope count (Supplementary Fig. 6-7) for each ethnicity was calculated as the summation of the epitope recognition potentials of all the epitopes identified by the HLA repertoire of that ethnicity.
4.4. Statistical analysis
In order to probe for any statistical differences between the average number of epitopes recognized by the individuals among the various super-populations, one way ANOVA test was conducted after grouping the ethnicities into super-populations. Additionally, t-test was also performed to test whether the average number of epitopes recognized in a super-population (and/or ethnicity) differed with respect to that of all other super-populations (and/or ethnicities) combined (Supplementary Table 6). The above tests were conducted both for the CD8 and CD4 epitopes individually as well as in combination. The p-values were corrected for multiple testing using Benjamini-Hochberg (BH) correction method. Further, a Principal Component Analysis (PCA) was performed to check the coherence of the antigen recognition potentials (in terms of the distribution of relevant HLA alleles) among the ethnicities (Fig. 3). The dudi.pca function from ade4 package was used for the purpose.
4.5. Epitope variations among SARS-CoV2 genomes
Nextstrain/augur pipeline (accessed on 21st April 2020 from https://github.com/nextstrain/ncov) was used to align 40,342 SARS-CoV2 genome sequences downloaded from GISAID using Wuhan-Hu-1/2019, as a reference sequence using MAFFT (Katoh and Standley, 2013). Conforming to the Nextstrain protocol, 130 nucleotides from 5′-end and 50 nucleotides from the 3′-end as well as single nucleotide positions 18,529, 29,849, 29,851, 29,853 were masked from multiple sequence alignment (MSA). An initial maximum likelihood (raw) phylogenetic tree (GTR model) was built using IQ-TREE (Nguyen et al., 2015) and further refined using default parameters in the pipeline. The tree was then processed to construct a TimeTree having the ancestor of the following two genomes, Wuhan-Hu-1/2019 and Wuhan/WH01/2019, at its root. Finally, using augur's translation step, a translated MSA of all 14 SARS-CoV2 proteins were retrieved. The 487 epitopic regions (w.r.t. reference genome Wuhan/Hu-1/2019) were cropped out from the translated MSA, and amino acid variations occurring in each epitope across 40,342 SARS-CoV2 isolates were obtained. A total of 4194 variants of ‘reference’ epitopes (VREs) could be identified from the 487 reference epitopes (see Table 1). Further, 4328 VREs for the 505 epitopes which could be recognized by the alleles encoded by any of the individuals (Samples) belonging to TGP were also obtained and used for the analysis depicted in Fig. 4.
4.6. Analysis of the genetic variability of SARS-CoV2 on immune recognition
To evaluate any possible effect of mutation occurring/ accumulating in the SARS-CoV2 genome over-time on immune recognition of the virus among different ethnicities, the mean epitope recognition potential for each individual belonging to an ethnic group was calculated with epitopes predicted from the first 10,000 and last 10,000 SARS-CoV2 genomes arranged in the chronological order of their collection dates (Fig. 4). For the purpose, the epitope recognition potential of each individual was computed as the sum of the number of CD8 and CD4 epitopes an individual can recognize using their HLA repertoire. Further, we also tried to estimate the variations of epitope recognition potential within each ethnic group for all the 40,342 variants of SARS-CoV2 genomes (Supplementary Fig. 8), where the mean epitope recognition potential of the ethnicities for each genome was calculated and arranged in the chronological order of their collection dates (in months). In this case, the epitope recognition potential of an ethnicity for a SARS-CoV2 variant was computed as was mentioned earlier (Supplementary Fig. 6).
Author contributions
TB, AD and SSM conceptualized the work and designed the protocol. NP, NKP and SB performed all the analyses. TB, NP, and AD wrote the manuscript. SSM supervised the project and reviewed the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgement
The presented work is based on SARS-CoV2 genome sequence data retrieved from the GISAID repository (www.gisaid.org). We also gratefully acknowledge the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative (Supplementary File 2). All submitters of data may be contacted directly via GISAID.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2021.198579.
Appendix. Supplementary materials
References
- Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W., Makhawi A.M. Design of a Multiepitope-Based Peptide Vaccine against the E Protein of Human COVID-19: an Immunoinformatics Approach [WWW Document] Biomed Res Int. 2020 doi: 10.1155/2020/2683286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L., Gouret P., Yeh J.-.H., Cristofaro J.D., Pontarotti P., Picard C., Paganini J. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0206512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso A., Magistroni P., Vespasiano F., Bella A., Bellino S., Puoti F., Alizzi S., Vaisitti T., Boros S., Grossi P.A., Trapani S., Lombardini L., Pezzotti P., Deaglio S., Brusaferro S., Cardillo M. Italian Network of Regional Transplant Coordinating Centers, 2021. HLA and AB0 Polymorphisms May Influence SARS-CoV-2 Infection and COVID-19 Severity. Transplantation. 2021;105:193–200. doi: 10.1097/TP.0000000000003507. [DOI] [PubMed] [Google Scholar]
- Ayukekbong J.A., Ntemgwa M.L., Ayukekbong S.A., Ashu E.E., Agbor T.A. COVID-19 compared to other epidemic coronavirus diseases and the flu. World J Clin Infect Dis. 2020;10:1–13. doi: 10.5495/wjcid.v10.i1.1. [DOI] [Google Scholar]
- Barquera R., Collen E., Di D., Buhler S., Teixeira J., Llamas B., Nunes J.M., Sanchez-Mazas A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020;96:277–298. doi: 10.1111/tan.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield S.F., Rook G.A., Scott E.A., Shanahan F., Stanwell-Smith R., Turner P. Time to abandon the hygiene hypothesis: new perspectives on allergic disease, the human microbiome, infectious disease prevention and the role of targeted hygiene. Perspect Public Health. 2016;136:213–224. doi: 10.1177/1757913916650225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciapuoti S., De Rosa A., Gelzo M., Megna M., Raia M., Pinchera B., Pontarelli A., Scotto R., Scala E., Scarano F., Scalia G., Castaldo G., Fabbrocini G., Gentile I., Parrella R. Immunocytometric analysis of COVID patients: a contribution to personalized therapy? Life Sci. 2020;261 doi: 10.1016/j.lfs.2020.118355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Hiyoshi A., Montgomery S. COVID-19 case-fatality rate and demographic and socioeconomic influencers: worldwide spatial regression analysis based on country-level data. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2020-043560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Coronavirus Disease 2019 (COVID-19) [WWW Document] Centers for Disease Control and Prevention. URL. 2020 https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/evidence-table.html (accessed 10.26.20) [Google Scholar]
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372 doi: 10.1136/bmj.n579. n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee B., Karandikar R.L., Mande S.C. Mortality due to COVID-19 in different countries is associated with their demographic character and prevalence of autoimmunity. Curr. Sci. 2021;120 8. [Google Scholar]
- Chen H.-.Z., Tang L.-.L., Yu X.-.L., Zhou J., Chang Y.-.F., Wu X. Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2. Infect Dis Poverty. 2020;88(9) doi: 10.1186/s40249-020-00713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correale P., Mutti L., Pentimalli F., Baglio G., Saladino R.E., Sileri P., Giordano A. HLA-B*44 and C*01 Prevalence Correlates with Covid19 Spreading across Italy. Int. J. Mol. Sci. 2020;21:E5205. doi: 10.3390/ijms21155205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio N., Walker C.R., Turakhia Y., Lanfear R., Corbett-Detig R., Goldman N. Mutation Rates and Selection on Synonymous Mutations in SARS-CoV-2. Genome Biol Evol. 2021;13 doi: 10.1093/gbe/evab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Chu Z., Yu F., Zha Y. Contriving Multi-Epitope Subunit of Vaccine for COVID-19: immunoinformatics Approaches. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: gISAID’s innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F.C., Schooley R.T. Treatment of COVID-19 – Evidence-Based or Personalized Medicine? Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vidal C., Moreno-García E., Hernández-Meneses M., Puerta-Alcalde P., Chumbita M., Garcia-Pouton N., Linares L., Rico V., Cardozo C., Martínez J.A., García F., Mensa J., Castro P., Nicolás J.M., Muñoz J., Vidal D., Soriano A. COVID19-Researchers, 2020. Personalized therapy approach for hospitalized patients with COVID-19. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciaa964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza F.F., McCabe A., Santos E.J.M., dos, Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A., Middleton D., Jones A.R. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48:D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D., Evans S.J., Walker A.J., Hickman G., Nightingale E., Schultze A., Rentsch C.T., Bates C., Cockburn J., Curtis H.J., Morton C.E., Bacon S., Davy S., Wong A.Y., Mehrkar A., Tomlinson L., Douglas I.J., Mathur R., Blomquist P., MacKenna B., Ingelsby P., Croker R., Parry J., Hester F., Harper S., DeVito N.J., Hulme W., Tazare J., Goldacre B., Smeeth L., Eggo R.M. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Increased risk of severe clinical course of COVID-19 in carriers of HLA-C*04:01 - EClinicalMedicine [WWW Document], n.d. URL https://www.thelancet.com/journals/eclinm/article/PIIS2589-5370(21)00379-5/fulltext (accessed 9.6.21). 2021. [DOI] [PMC free article] [PubMed]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic. Acids. Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor S.-.S., Omae Y., Nishida N., Sugiyama M., Kinoshita N., Suzuki T., Suzuki M., Suzuki S., Izumi S., Hojo M., Ohmagari N., Mizokami M., Tokunaga K. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, Age and Sex Are Associated With Severity of Japanese COVID-19 With Respiratory Failure. Front Immunol. 2021;12:1134. doi: 10.3389/fimmu.2021.658570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichel B., Falke S., Hilgenfeld R., Redecke L., Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7–10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Coronavirus Disease 2019 (COVID-19) 2020:23–31. doi: 10.1007/978-981-15-4814-7_3. [DOI] [Google Scholar]
- Langton D.J., Bourke S.C., Lie B.A., Reiff G., Natu S., Darlay R., Burn J., Echevarria C. The influence of HLA genotype on the severity of COVID-19 infection. HLA. 2021;98:14–22. doi: 10.1111/tan.14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020:1–8. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yuan L., Dai G., Chen R.A., Liu D.X., Fung T.S. Regulation of the ER Stress Response by the Ion Channel Activity of the Infectious Bronchitis Coronavirus Envelope Protein Modulates Virion Release, Apoptosis, Viral Fitness, and Pathogenesis. Front Microbiol. 2020;10 doi: 10.3389/fmicb.2019.03022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Ting S., Yufei H., Wendong L., Yubo F., Jing Z. Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova, A., Kudlay, D., Kudryavtsev, I., Starshinova, A., Yablonskiy, P., Shoenfeld, Y., 2021. Immunogenetic Predictors of Severe COVID-19. Vaccines (Basel) 9, 211. 10.3390/vaccines9030211. [DOI] [PMC free article] [PubMed]
- Mercatelli D., Giorgi F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini F., Torsiello E., Spiezia F., Oliva F., Tingart M., Maffulli N. Association between HLA genotypes and COVID-19 susceptibility, severity and progression: a comprehensive review of the literature. Eur. J. Med. Res. 2021;26:84. doi: 10.1186/s40001-021-00563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N., McMichael A. Antigen presentation in virus infection. Curr. Opin. Immunol. 1992;4:401–407. doi: 10.1016/S0952-7915(06)80030-0. [DOI] [PubMed] [Google Scholar]
- Naz A., Shahid F., Butt T.T., Awan F.M., Ali A., Malik A. Designing Multi-Epitope Vaccines to Combat Emerging Coronavirus Disease 2019 (COVID-19) by Employing Immuno-Informatics Approach. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli A., Andreani M., Biancolella M., Liberatoscioli L., Passarelli C., Colona V.L., Rogliani P., Leonardis F., Campana A., Carsetti R., Andreoni M., Bernardini S., Novelli G., Locatelli F. HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA. 2020;96:610–614. doi: 10.1111/tan.14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., Yeats C., du Plessis L., Maloney D., Medd N., Attwood S.W., Aanensen D.M., Holmes E.C., Pybus O.G., Rambaut A. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021 doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan R.K., Biswas M., Khandaker M.U. Time series prediction of COVID-19 by mutation rate analysis using recurrent neural network-based LSTM model. Chaos Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.110018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Sidney J., Sette A., Peters B. TepiTool: a Pipeline for Computational Prediction of T Cell Epitope Candidates. Curr Protoc Immunol. 2016;114 doi: 10.1002/cpim.12. 18.19.1-18.19.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C., Grimes J.M., Antson A.A., Bayfield O.W., Hawkins D.E.D.P., Ker D.-.S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-.L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B.M., Fry J.W., Schwabe N.F., Semple M.G., Baillie J.K., Moore S.C., Openshaw P.J.M., Ansari M.A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-.P., Cornall R.J., Conlon C.P., Klenerman P., Screaton G.R., Mongkolsapaya J., McMichael A., Knight J.C., Ogg G., Dong T. Broad and strong memory CD4 + and CD8 + T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanti S., Deelen J., Gallina A.M., Caputo M., Citro M., Abate M., Sacchi N., Vecchione C., Martinelli R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid-19. J. Transl. Med. 2020;18 doi: 10.1186/s12967-020-02515-5. 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic. Acids. Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López J.P., Carnalla-Cortés M., Pacheco-Olvera D.L., Ocampo-Godínez J.M., Oliva-Ramírez J., Moreno-Manjón J., Bernal-Alferes B., López-Olmedo N., García-Latorre E., Domínguez-López M.L., Reyes-Sandoval A., Jiménez-Zamudio L. A bioinformatic prediction of antigen presentation from SARS-CoV-2 spike protein revealed a theoretical correlation of HLA-DRB1×01 with COVID-19 fatality in Mexican population: an ecological approach. J. Med. Virol. 2021;93:2029–2038. doi: 10.1002/jmv.26561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., Raghava, G.P.S., 2004. BcePred: Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using Physico-Chemical Properties, in: Nicosia, G., Cutello, V., Bentley, P.J., Timmis, J. (Eds.), Artificial Immune Systems, Lecture Notes in Computer Science. Springer, Berlin, Heidelberg, pp. 197–204. 10.1007/978-3-540-30220-9_16.
- Satarker S., Nampoothiri M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020;51:482. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Ostrov D.A., Rananaware S., Jain P.K., Nguyen C.Q. 2021. Identification of Risk and Protective Human Leukocyte Antigens in COVID-19 Using Genotyping and Structural Modeling. [DOI] [Google Scholar]
- Shkurnikov M., Nersisyan S., Jankevic T., Galatenko A., Gordeev I., Vechorko V., Tonevitsky A. Association of HLA Class I Genotypes With Severity of Coronavirus Disease-19. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.641900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanina H., Madhugiri R., Bylapudi G., Schultheiß K., Karl N., Gulyaeva A., Gorbalenya A.E., Linne U., Ziebuhr J. Coronavirus replication–transcription complex: vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. PNAS. 2021;118 doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Delaney M., Shah R.K., Campos J.M., Wessel D.L., DeBiasi R.L. Comparison of Clinical Features of COVID-19 vs Seasonal Influenza A and B in US Children. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.20495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorci G., Faivre B., Morand S. Explaining among-country variation in COVID-19 case fatality rate. Sci Rep. 2020;10:18909. doi: 10.1038/s41598-020-75848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- topuzoğullari m., acar t., pelit arayici p., uçar b., uğurel e., abamor e.ş, arasoğlu t., turgut-balik d., derman s. An insight into the epitope-based peptide vaccine design strategy and studies against COVID-19. Turk J Biol. 2020;44:215–227. doi: 10.3906/biy-2006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020:1–8. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfoort N., van der Aa E., Woltman A.M. Understanding MHC Class I Presentation of Viral Antigens by Human Dendritic Cells as a Basis for Rational Design of Therapeutic Vaccines. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-.W., Lin Y.-.C., Pai T.-.W., Chang H.-.T. Prediction of B-cell Linear Epitopes with a Combination of Support Vector Machine Classification and Amino Acid Propensity Identification [WWW Document] Journal of Biomedicine and Biotechnology. 2011 doi: 10.1155/2011/432830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang Xiaobao, Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang Xiuna, Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus Disease (COVID-19) Dashboard [WWW Document], n.d. URL https://covid19.who.int (accessed 10.26.20).
- WHO | Variant analysis of SARS-CoV-2 genomes [WWW Document], n.d. WHO. 10.2471/BLT.20.253591. [DOI]
- Wilson E.A., Hirneise G., Singharoy A., Anderson K.S. Total predicted MHC-I epitope load is inversely associated with population mortality from SARS-CoV-2. Cell Reports Medicine. 2021;2 doi: 10.1016/j.xcrm.2021.100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: sARS-CoV-2 and SARS-CoV. Viruses. 2020;12 doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.-.H., Ding Y.-.B., Xu Z., Pu R., Li P., Yan J., Liu J.-.L., Meng F.-.P., Huang L., Shi L., Jiang T.-.J., Qin E.-.Q., Zhao M., Zhang D.-.W., Zhao P., Yu L.-.X., Wang Z.-.H., Hong Z.-.X., Xiao Z.-.H., Xi Q., Zhao D.-.X., Yu P., Zhu C.-.Z., Chen Z., Zhang S.-.G., Ji J.-.S., Wang F.-.S., Cao G.-.W. Increased circulating level of interleukin-6 and CD8+ T cell exhaustion are associated with progression of COVID-19. Infect Dis Poverty. 2020;9:161. doi: 10.1186/s40249-020-00780-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer T., Waseem M., Waqar W., Dar H.A., Shehroz M., Naz K., Ishaq Z., Ahmad T., Ullah N., Bakhtiar S.M., Muhammad S.A., Ali A. 2020. Anti-COVID-19 Multi-Epitope Vaccine Designs Employing Global Viral Genome sequences. PeerJ 8, e9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lund O., Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics. 2009;25:1293–1299. doi: 10.1093/bioinformatics/btp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.