Figure 4. Schematic of autoSTOMP mediated targeting of discrete IgG4+ lesions across multiple esophageal biopsy sections.
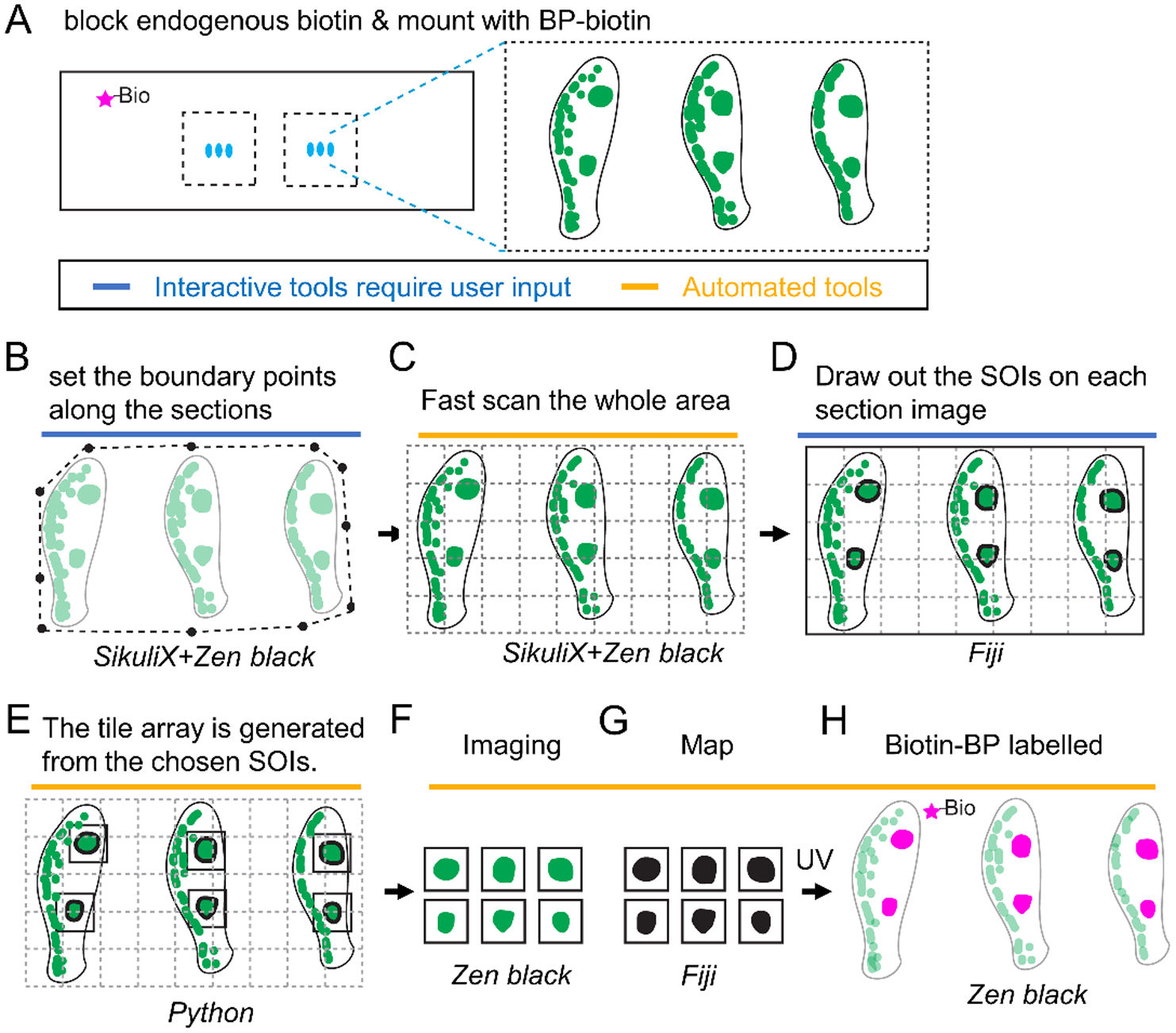
1 mm esophagus punch biopsies are isolated from a patient with active Eosinophilic Esophagitis (EoE). A, 6–8 sections per slide are stained for IgG4 (green), endogenous biotins are blocked and mounted with BP-biotin. A typical section contains 5 tiles at 340 μm × 340 μm per field of view using a 25x magnification objective lens. B–H, The SikuliX script allows the user to set the boundary points for all sections in Zen Black (B). C, The boundary points then direct low resolution tile scan in Zen Black. D, IgG4+ structures of interest (SOI) are identified by the user on each section in Fiji. E, A Python script maps the coordinates of each SOI to generate a tile array of SOI-containing fields of view. F–H, A SikuliX script automates SOI imaging in Zen black (F, green), MAP file generation in Fiji (G, black) and BP-biotin crosslinking in Zen Black (H, magenta) as described in Figure 1.
