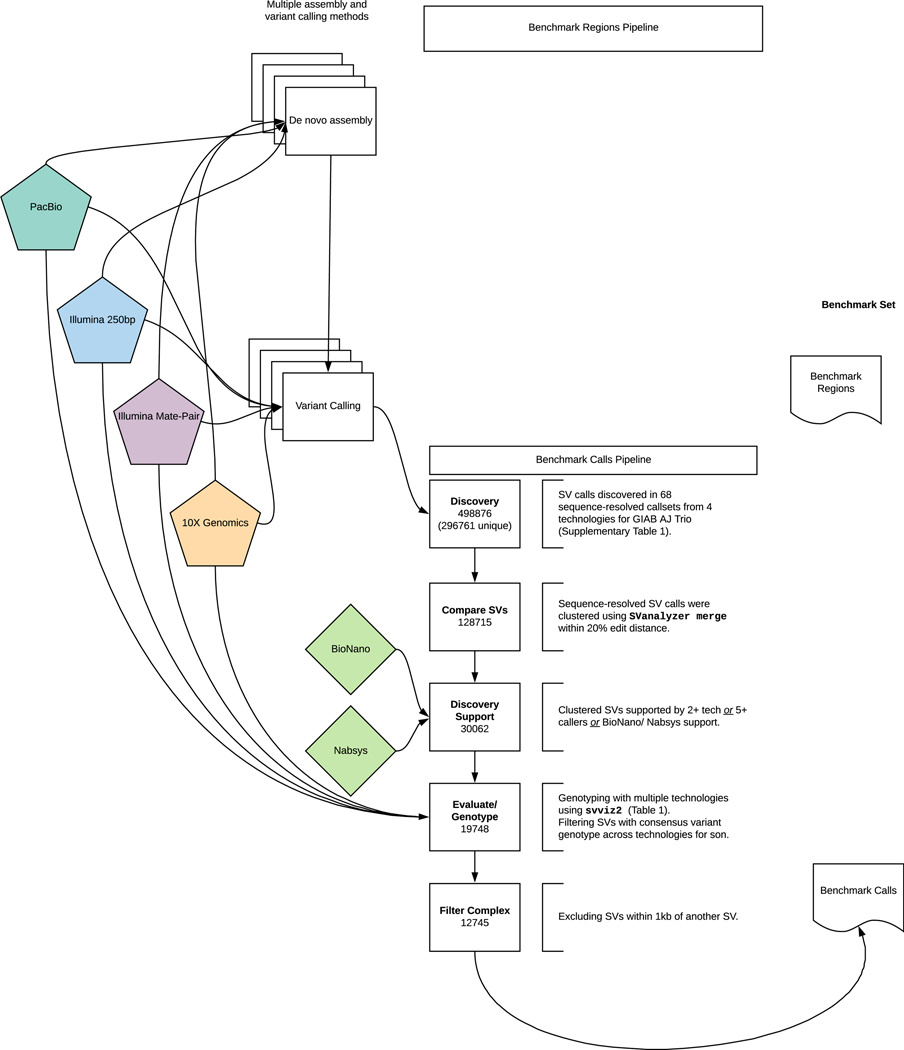
Figure 2: Process to integrate SV callsets and diploid assemblies from different technologies and analysis methods and form the benchmark set.
The input datasets are depicted in the center of the figure with the benchmark calls and region pipelines to the left and right of the input data, respectively. The number of variants in each step of the benchmark calls integration pipeline is indicated in the white boxes. See the Methods section for additional description of the pipeline steps. Briefly, approximately 0.5 million input SV calls were locally clustered based on their estimated sequence change, and we kept only those discovered by at least two technologies or at least 5 callsets in the trio. We then used svviz with short, linked, and long reads to evaluate and genotype these calls, keeping only those with a consensus heterozygous or homozygous variant genotype in the son. We filtered potentially complex calls in regions with multiple discordant SV calls, as well as regions around 20 bp to 49 bp indels, and our final Tier 1 benchmark set included 12745 total insertions and deletions ≥50 with 9357 inside the 2.51 Gbp of the genome where diploid assemblies had no additional SVs beyond those in our benchmark set. We also define a Tier 2 set of 6007 additional regions where there was substantial support for one or more SVs but the precise SV was not yet determined.