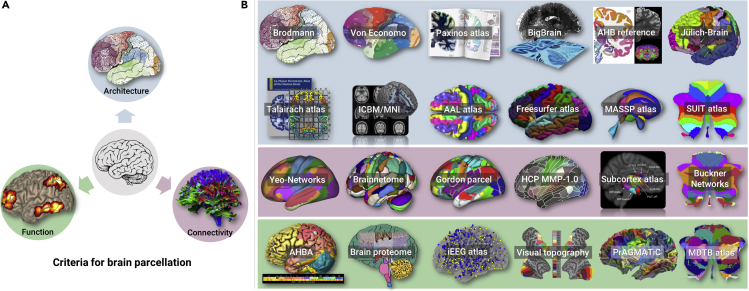
Figure 1.
Parcellation Criteria and Typical Human Brain Atlases
Criteria for human brain parcellation (A), including architecture, function, and connectivity, and the collections of available human brain atlases (B). As one of the criteria for brain parcellation, function contains diverse and complex contents, so here we list information including genetic and protein expressions in this category. The available human brain atlases are listed alphabetically as follows:
AAL atlas: automated anatomical labeling atlas, https://www.gin.cnrs.fr/en/tools/aal/
AHBA: Allen Human Brain Atlas integrating anatomic and genomic information https://human.brain-map.org
AHB reference: Allen Human Brain reference atlas, a 2D coronal cellular resolution anatomical reference atlas, http://atlas.brain-map.org
BigBrain: microscopic 3D model with maps of cytoarchitectonic structures, https://bigbrainproject.org
Brain proteome: protein-coding genes are classified based on RNA expression in the brain https://www.proteinatlas.org/humanproteome/brain
Brainnetome: human Brainnetome atlas based on connectional architecture http://atlas.brainnetome.org
Buckner-networks: cerebellar parcellation estimated by intrinsic functional connectivity http://surfer.nmr.mgh.harvard.edu/fswiki/CerebellumParcellation_Buckner2011
Caret surface: http://brainvis.wustl.edu/wiki/index.php/Caret:About
Freesurfer atlas: Desikan-Killiany Atlas and Destrieux Atlas https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation
Gordon parcel: resting-state functional connectivity-boundary map-derived parcels https://sites.wustl.edu/petersenschlaggarlab/resources/
HCP MMP-1.0: a multimodal parcellation of the human cerebral cortex, https://balsa.wustl.edu/study/show/RVVG
ICBM/MNI: an unbiased standard MRI brain template, http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009
iEEG Atlas: Montreal Neurological Institute (MNI) Open Intracranial EEG Atlas, https://mni-open-ieegatlas.research.mcgill.ca
Jülich-Brain: a 3D probabilistic atlas of the human brain's cytoarchitecture, https://jubrain.fz-juelich.de/apps/cytoviewer2/cytoviewer.php##mitte
https://ebrains.eu/services/atlases/brain-atlases
MDTB atlas: multi-domain task battery maps http://www.diedrichsenlab.org/imaging/mdtb.htm
Paxinos atlas: the fourth edition of Atlas of the Human Brain, http://atlas.thehumanbrain.info
PrAGMATiC: A Probabilistic and Generative Model of Areas Tiling the Cortex, https://gallantlab.org/huth2016
Subcortex parcel: Melbourne subcortex atlas, https://github.com/yetianmed/subcortex
SUIT atlas: a spatially unbiased atlas template of the human cerebellum, http://www.diedrichsenlab.org/imaging/suit.htm
Talairach atlas: a 3D coordinate system of Talairach space, http://www.talairach.org
Visual topography: probabilistic maps of visual topography in human cortex https://scholar.princeton.edu/napl/resources
Yeo-Networks: cortical parcellation estimated by intrinsic functional connectivity, https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation_Yeo2011