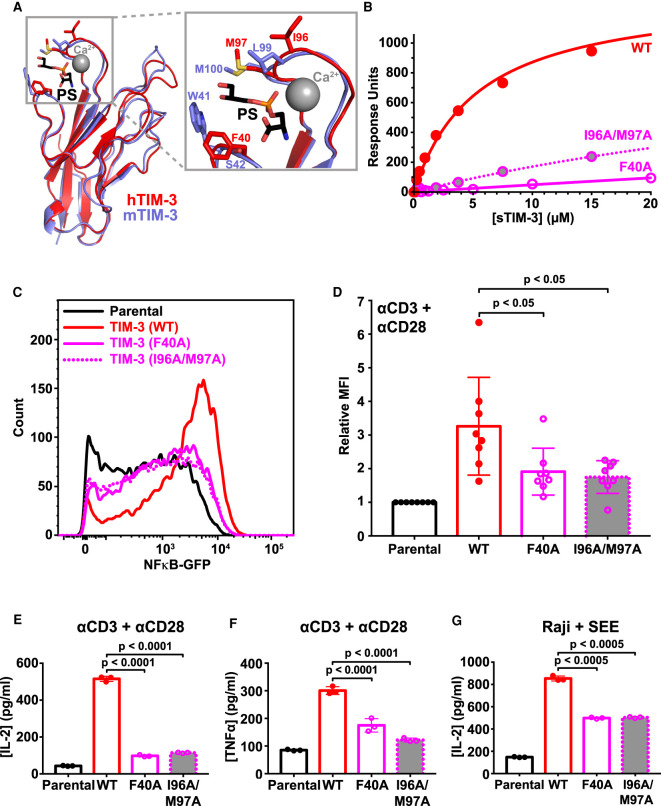
Figure 5. Mutated TIM-3 variants with impaired PS-binding lose co-stimulatory signaling.
(A) Details of the PS-binding site in murine TIM-3 (PDB: 3KAA — blue) overlaid on human TIM-3 (PDB: 6TXZ — red), revealing key interacting residues [34,82]. Inset at the right shows the PS-binding pocket, where short-chain PS is sandwiched by L99/M100 and W41 in mTIM-3 (I96/M97 and F40 in human TIM-3). A calcium ion (gray sphere) also sits in the binding pocket and interacts with the negatively charged PS headgroup. (B) PS binding of wild-type sTIM-3 (solid red curve), sTIM-3F40A (solid magenta curve), and sTIM-3I96A/M97A (dotted magenta curve) were analyzed using SPR by flowing the purified sTIM-3 variants over 20% DOPS/80% DOPC lipid vesicles immobilized on an L1 chip (in the presence of 1 mM CaCl2). Binding curves are representative of at least three independent experiments. Kd values for sTIM-3F40A and sTIM-3I96A/M97A were too high to measure, both appearing to exceed ∼200 μM (although PS binding was detectable). (C) A GFP reporter was used to measure NF-κB transcriptional activity downstream of TCR activation with 1 μg/ml αCD3 plus 0.5 μg/ml αCD28 for 16 h. Representative histogram of NF-κB-driven GFP expression is shown for parental NF-κB GFP reporter cells (black), and those expressing TIM-3WT (solid red curve), TIM-3F40A (solid magenta curve), and TIM-3I96A/M97A (dotted magenta curve) variants. (D) Relative mean GFP fluorescence intensity (MFI), expressed as fold change over parental cells within each experiment, was determined across eight biological replicates for parental NF-κB GFP reporter cells (black) or those expressing TIM-3WT (solid red line, open bar), TIM-3F40A (solid magenta line, open bar and data points), or TIM-3I96A/M97A (dotted magenta line, gray filled bar and data points). Means ± SD are plotted, with P-values determined by two-tailed, unpaired Student's t-tests. (E) ELISA analysis of IL-2 secreted into the cell culture medium by parental NF-κB GFP reporter cells, and those expressing, TIM-3WT (solid red line, open bar), TIM-3F40A (solid magenta line, open bar and data points), or TIM-3I96A/M97A (dotted magenta line, gray filled bar and data points) following stimulation by αCD3/αCD28, as described in (C). Means ± SD are plotted for three biological repeats. P-values comparing TIM-3WT with the mutated TIM-3 variants were determined using two-tailed, unpaired Student's t-tests. (F) Analysis of TNFα secretion into the cell culture medium was performed using a Luminex assay. Cells were stimulated with αCD3/αCD28 (1 μg/ml each) for 16 h. Mean values ± SD are plotted for parental (black line, open bar), TIM-3WT (solid red line, open bar), TIM-3F40A (solid magenta line, open bar and points), and TIM-3I96A/M97A (dotted magenta line, gray filled bar and data points). Two-tailed, unpaired Student's t-tests were used to compare TIM-3WT with mutated TIM-3 variants for three biological repeats. (G) SEE-loaded Raji B cells were used to stimulate NF-κB GFP reporter Jurkat cells to recapitulate the formation of an immunological synapse. IL-2 production following Raji B cell stimulation was determined by ELISA analysis for parental NF-κB GFP reporter cells (solid black line, open bar) and cells expressing TIM-3WT (solid red line, open bar), TIM-3F40A (solid magenta line, open bar and data points), or TIM-3I96A/M97A (dotted magenta line, gray filled bar and data points). Mean ± SD are plotted for three biological repeats, with P-values comparing TIM-3WT with the mutated TIM-3 variants determined using two-tailed, unpaired Student's t-tests.