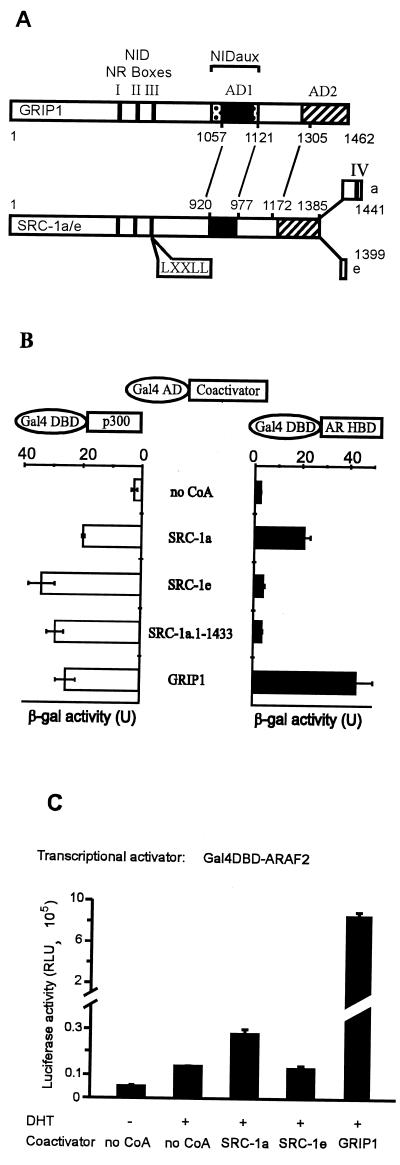
FIG. 1.
Binding of GRIP1, SRC-1a, and SRC-1e to AR HBD correlates with coactivator effect on AR AF-2. (A) Alignment and functional domains of p160 coactivators. NID, NR HBD interaction domain; vertical solid bars, NR boxes I, II, and III (LXXLL motifs); NIDaux (spotted box), auxiliary domain in GRIP1 required in addition to NR boxes for efficient binding of AR HBD but not TR HBD; AD1 (solid box) and AD2 (striped box), two autonomous activation domains; numbers, amino acids of GRIP1 and SRC-1. The lines between GRIP1 and SRC-1 amino acid numbers indicate alignment of homologous regions. NIDaux and AD1 overlap within GRIP1 amino acids 1011 and 1121; SRC-1 lacks NIDaux and SRC-1a contains a fourth NR box (IV). (B) Interaction of p160 coactivators with AR HBD in yeast two-hybrid assays. Gal4 AD, fused to the indicated coactivator, was expressed in yeast strain SFY526 along with a Gal4DBD-ARHBD or Gal4DBD-p300 fusion protein; when AR HBD was present, yeast cells were grown in 100 nM DHT. Activation of the integrated β-Gal reporter gene controlled by Gal4 binding sites was determined by measuring β-Gal activity in cell extracts. (C) Enhancement of AR AF-2 function by p160 coactivators. CV-1 cells were transfected with 0.5 μg of pM vector encoding Gal4DBD-ARAF2, 0.5 μg of GK1 reporter gene, and 1.0 μg of pSG5.HA (no CoA) or the same vector encoding the indicated coactivator. Luciferase activity, shown in relative light units (RLU), observed with or without DHT treatment is shown.