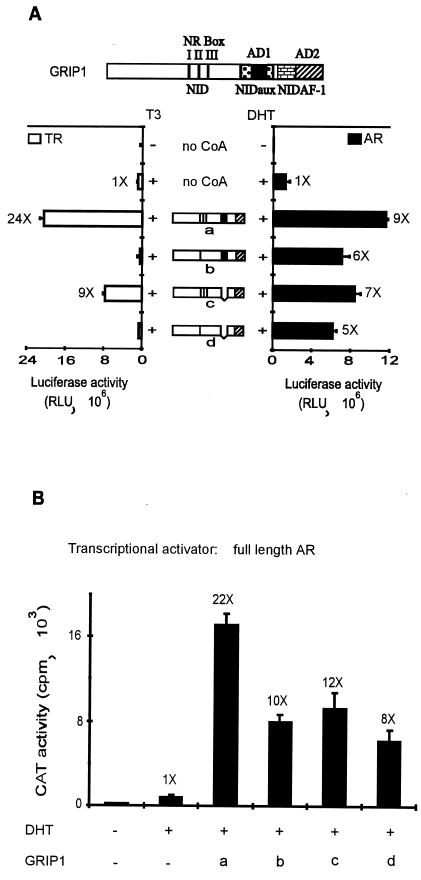
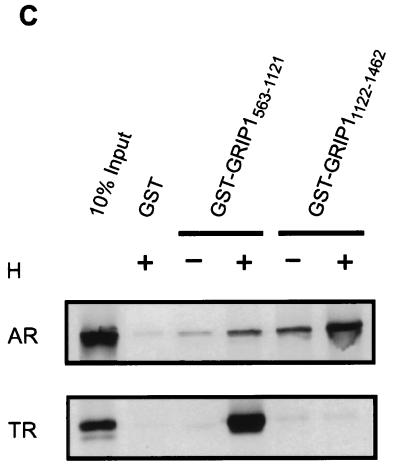
FIG. 8.
Differential use of NR-binding regions and ADs of GRIP1 by AR and TR. (A) Coactivator function of GRIP1 mutants with AR and TR. CV-1 cells were transiently transfected with 0.25 μg of pSVAR0 or pCMX.hTRβ1, 0.5 μg of reporter gene MMTV-LUC (for AR) or MTV(TRE)-LUC (for TR), and 1.25 μg of pSG5.HA vector encoding no coactivator (no CoA) or full-length GRIP1 with a wild-type sequence (a) or with mutations in the NR boxes II and III (b), deletion of AD1 (c), or both mutations (d). Luciferase activity was determined from extracts of cells grown without or with 20 nM DHT (for AR) or 20 nM T3 (for TR) as indicated. Functional domains in diagrams are as in Fig. 1A; in addition, the brick pattern represents NIDAF-1. Numbers beside data bars indicate the activity relative to that with AR or TR in the presence of hormone and absence of GRIP1. (B) Coactivator function of GRIP1 mutants with AR tested on a rat probasin promoter. CV-1 cells were transiently transfected with 0.25 μg of pSVAR0, 0.5 μg of rPB-CAT reporter gene, and pSG5.HA vectors encoding wild-type or mutant GRIP1; letters a to d represent GRIP1 protein species as in panel A. (C) Binding of GRIP1 N- or C-terminal fragments to AR and TR in vitro. 35S-labeled full-length AR or TR was synthesized in vitro and then incubated with glutathione-Sepharose-bound GST or GST-GRIP1 fusion proteins in the absence or presence of the appropriate hormone (H, 1 μM DHT for AR or 1 μM T3 for TR) as indicated. Bound proteins were eluted and analyzed by SDS-PAGE and autoradiography.