Visual Abstract
Keywords: membranous nephropathy, PLA2R, HLA, glomerular disease, genetics, haplotypes, prognosis
Abstract
Background and objectives
Associations between HLA alleles and susceptibility to M-type phospholipase A2 receptor (PLA2R)–related membranous nephropathy have been well defined previously in Chinese patients. However, the relationships between HLA alleles and kidney outcome remain unclear.
Design, setting, participants, & measurements
Five HLA genes (DRB1, DQA1, DQB1, DRB3, and DRB5) were genotyped in a prospective cohort of 392 patients with PLA2R-related membranous nephropathy. The associations between HLA alleles and kidney outcomes were studied.
Results
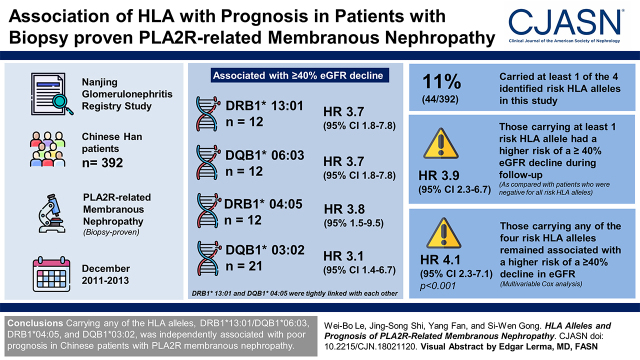
A total of 79 HLA alleles were identified in this study. Four HLA alleles, DRB1*13:01 (n=12; hazard ratio, 3.7; 95% confidence interval, 1.8 to 7.8; P<0.001), DQB1*06:03 (n=12; hazard ratio, 3.7; 95% confidence interval, 1.8 to 7.8; P<0.001), DRB1*04:05 (n=12; hazard ratio, 3.8; 95% confidence interval, 1.5 to 9.5; P=0.004), and DQB1*03:02 (n=21; hazard ratio, 3.1; 95% confidence interval, 1.4 to 6.7; P=0.005), were associated with a ≥40% eGFR decline during follow-up. DRB1*13:01 and DQB1*06:03 were tightly linked with each other. Forty-four of the 392 patients (11%) carried at least one of the four identified risk HLA alleles in this study. Compared with patients who were negative for all risk HLA alleles, those carrying at least one risk HLA allele had a significant risk of a ≥40% eGFR decline during follow-up (hazard ratio, 3.9; 95% confidence interval, 2.3 to 6.7; P<0.001). After adjusting for age, sex, proteinuria, albumin, eGFR, and anti-PLA2R antibody levels, multivariable Cox analysis showed that patients carrying any of the four risk HLA alleles remained associated with a higher risk of a ≥40% decline in eGFR (hazard ratio, 4.1; 95% confidence interval, 2.3 to 7.1; P<0.001).
Conclusions
Carrying any of the HLA alleles, DRB1*13:01/DQB1*06:03, DRB1*04:05, and DQB1*03:02, was independently associated with poor prognosis in Chinese patients with PLA2R-related membranous nephropathy.
Introduction
Membranous nephropathy is the leading cause of nephrotic syndrome. Kidney biopsy registry data have shown a rising incidence of membranous nephropathy in China during the last decade (1,2). In a landmark study in 2009 by Beck et al. (3), M-type phospholipase A2 receptor (PLA2R) was identified as a major antigen in idiopathic membranous nephropathy. Approximately 70%–80% of patients presumed to have idiopathic membranous nephropathy have a positive test for anti-PLA2R antibody at the time of kidney biopsy diagnosis of membranous nephropathy, and 3%–5% have circulating antibodies to thrombospondin type-1 domain–containing 7A or neural epidermal growth factor–like 1 protein (4–8). The susceptibility to idiopathic membranous nephropathy is strongly associated with genetic factors, including HLA alleles and PLA2R variants (9–16). A genome-wide association study in White individuals identified strong associations between the HLA region and the PLA2R1 locus encoding the dominant antigen of membranous nephropathy (10). The HLA associations may differ between East Asians and Europeans. DRB1*15:01 (DRB1*15:01-DQA1*01:02-DQB1*06:02-DRB5*01:01 haplotype) and DRB3*02:02 (DRB3*02:02-DRB1*03:01-DQB1*02:01-DQA1*05:01 haplotype) are the major independent susceptibility alleles in East Asians, whereas DQA1*05:01 is the strongest allele in Europeans (4,11,17).
Because spontaneous remission is common in membranous nephropathy and immunosuppressive agents have appreciable toxicity, initial therapy for patients with membranous nephropathy is supportive; immunosuppressive therapy is recommended for patients with a high risk of developing progressive kidney failure (18). The major risk factors for kidney failure in patients with membranous nephropathy include older age at onset, persistent nephrotic-range proteinuria, lower eGFR, progressive decline in kidney function, and higher levels of anti-PLA2R antibodies (19). Most patients need to wait for at least 6 months to assess their risk of progressive loss of kidney function. Therefore, novel biomarkers are urgently required to identify high-risk patients with disease progression. The associations between HLA alleles and susceptibility to idiopathic membranous nephropathy have already been well studied. However, whether HLA alleles could be associated with kidney prognosis in membranous nephropathy remains poorly defined (20). This study aimed to address this issue in a prospective cohort of 392 Chinese patients with PLA2R-related membranous nephropathy.
Materials and Methods
Participants and Samples: Histocompatibility Leukocyte Antigen Typing
The details of the participants and HLA typing methods have been described in previous publications (5,9) and are briefly summarized here. A total of 392 Chinese Han patients who were diagnosed with biopsy-proven PLA2R-related membranous nephropathy between December 2011 and December 2013 were enrolled from the Nanjing Glomerulonephritis Registry Study. All patients were clinically ruled out for secondary membranous nephropathy. Patients positive for serum anti-PLA2R antibodies (>20 RU/ml) were defined as having PLA2R-related membranous nephropathy in this study. Patients were followed up by nephrologists at the outpatient department of the National Clinical Research Center of Kidney Diseases. A total of 14 patients were lost to follow-up after kidney biopsy. Medical records were collected at the time of kidney biopsy and during a median follow-up time of 63 (interquartile range [IQR], 11–84) months. All follow-up data were updated to January 2020. For the 392 patients enrolled in this study, there were a total of 6466 measurements of eGFR during the follow-up period. eGFR measurements were taken a median of 16 (IQR, 8–22) times for each patient during the follow-up.
Serum anti-PLA2R antibody levels were detected by ELISA. The serum creatinine level was measured with enzymatic assays. All of the HLA typing was performed by BGI-Shenzhen. Two methods of HLA typing were used in this study. Ninety-nine patients were captured by an HLA array that contained the entire 5-Mb MHC region (9). HLA target sequencing was performed on the HiSEquation 2000 platform to generate 100-bp paired-end reads. HLA typing in the discovery cohort was performed by SOAP-HLA Software (9,21). The remaining 293 patients were genotyped using the Sanger platform for the five selected HLA genes (HLA-DRB1, HLA-DQB1, HLA-DQA1, HLA-DRB3, and HLA-DRB5). The research protocol was approved by the Institutional Review Board of Jinling Hospital, Nanjing University and conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants. All human kidney and blood specimens were obtained from the Biobank of the National Clinical Research Center of Kidney Diseases at Jinling Hospital. A total of 385 ethnically matched healthy control samples were also included from the biobank.
Statistical Analyses
The kidney outcome event was a ≥40% eGFR decline defined as the time to the first occurrence of ≥40% eGFR decline from baseline during follow-up. The association for every HLA allele was analyzed by a set of biallelic tests, in which each allele was marked as presence or absence. The eGFR was estimated using the Chronic Kidney Disease Epidemiology Collaboration equation (21). Associations between HLA alleles and kidney outcome were analyzed by Cox proportional hazards models. Alleles with a significance level <0.01 were considered in further analysis. The time from kidney biopsy to kidney outcome event (≥40% eGFR decline) was also analyzed using Kaplan–Meier curve analysis.
All 392 patients were included in the statistical analysis, and there were no missing data in this study. Normally distributed variables are expressed as the mean ± SD. Nonparametric variables are expressed as the median and range. Categorical variables are expressed in percentages. Two-tailed P=0.01 findings were considered statistically significant. To calculate the pairwise linkage disequilibrium (LD) between DRB1*13:01 and DQB1*06:03, the carrying states of DRB1*13:01 and DQB1*06:03 were marked as present or absent for each individual, and thus, the calculation of LD statistics between DRB1*13:01 and DQB1*06:03 was simplified into the case of two locus, two allele. Finally, the LD statistics of R2 and D' between DRB1*13:01 and DQB1*06:03 were estimated via the expectation-maximization algorithm in 777 individuals (392 patients and 385 healthy controls) using PLINK 1.07. All other statistical analyses were carried out using R (version 4.0.2).
Results
Associations between Histocompatibility Leukocyte Antigen Alleles and Kidney Outcome
The clinical characteristics of 392 Chinese patients with PLA2R-related membranous nephropathy are shown in Table 1. Five HLA genes (DRB1, DQA1, DQB1, DRB3, and DRB5) were genotyped, and a total of 79 HLA alleles were identified in these patients (Supplemental Table 1). Cox proportional hazards models were conducted to assess associations between HLA alleles and kidney outcomes. Among the 79 HLA alleles (Supplemental Table 1), two alleles, DRB1*13:01 and DQB1*06:03 (both hazard ratio [HR], 3.7; 95% confidence interval [95% CI], 1.8 to 7.8; P<0.001), were associated with a ≥40% eGFR decline during follow-up. There were 12 patients who carried DRB1*13:01 and DQB1*06:03. DRB1*13:01 and DQB1*06:03 were linked with each other in this study (R2=1; D'=1.0; n=777). All patients with PLA2R-related membranous nephropathy with DRB1*13:01 (n=12) were also positive for DQB1*06:03, and all patients without DRB1*13:01 (n=380) were also negative for DQB1*06:03.
Table 1.
Clinical characteristics of patients with M-type phospholipase A2 receptor–related membranous nephropathy with or without any HLA risk alleles
| Clinical Item | Overall | Any | None |
| Patients, n | 392 | 44 | 348 |
| Men | 242 (62%) | 28 (64%) | 214 (62%) |
| Age, yr | 44±16 | 44±15 | 44±16 |
| Proteinuria at baseline, g/24 h | 4.4±2.8 | 4.7±2.4 | 4.3±2.5 |
| Albumin at baseline, g/dl | 2.8±0.5 | 2.8±0.5 | 2.8±0.5 |
| Serum creatinine at baseline, mg/dl | 0.83±0.29 | 0.85±0.33 | 0.82±0.28 |
| eGFR at baseline, ml/min per 1.73 m2 | 102±26 | 101±29 | 102±25 |
| Anti-PLA2R antibodies at baseline, U/ml, median (IQR) | 81.6 (39.0–168.8) | 86.4 (45.9–152.0) | 79.2 (38.3–132.0) |
| eGFR at last follow-up, ml/min per 1.73 m2 | 90±38 | 71±46 | 92±36 |
HLA risk alleles: DRB1*13:01, DQB1*06:03, DRB1*04:05, or DQB1*03:02. PLA2R, M-type phospholipase A2 receptor; IQR, interquartile range.
Two other HLA alleles, DRB1*04:05 (HR, 3.8; 95% CI, 1.5 to 9.5; P=0.004) and DQB1*03:02 (HR, 3.1; 95% CI, 1.4 to 6.7; P=0.005), were marginally associated with a ≥40% eGFR decline. After adjusting for the DRB1*13:01, the association signals of DRB1*04:05 (HR, 4.2; 95% CI, 1.7 to 10.7; P=0.002) and DQB1*03:02 (HR, 3.4; 95% CI, 1.5 to 7.6; P=0.002) remained unchanged and enhanced slightly, indicating that DRB1*04:05 and DQB1*03:02 were risk alleles independent of DRB1*13:01/DQB1*06:03. When the three risk alleles were all included in the Cox model, the associations remained strong for all of them: DRB1*13:01/DQB1*06:03 (HR, 4.4; 95% CI, 2.1 to 9.4; P<0.001), DRB1*04:05 (HR, 4.5; 95% CI, 1.8 to 11.4; P=0.001), and DQB1*03:02 (HR, 3.6; 95% CI, 1.6 to 7.9; P=0.002). These results indicated that DRB1*13:01/DQB1*06:03, DRB1*04:05, and DQB1*03:02 were independent risk alleles associated with kidney outcome. There were 12 patients positive for DRB1*04:05 and 21 patients positive for DQB1*03:02. Of the 21 patients positive for DQB1*03:02, only one patient was positive for DRB1*04:05. Forty-four of the 392 patients (11%) carried at least one of the four identified risk HLA alleles in this study. Compared with 348 patients who were negative for all of the risk HLA alleles, those carrying at least one risk HLA allele had a significant risk of a ≥40% eGFR decline during follow-up (HR, 3.9; 95% CI, 2.3 to 6.7; P<0.001).
Although DRB1*15:01 and DRB3*02:02 (or DRB1*03:01) are independently associated with the development of idiopathic membranous nephropathy, neither of them is associated with kidney outcome. In this study, the DRB1*15:02 allele did not show any association with a higher risk of a ≥40% eGFR decline during follow-up. In contrast, patients with the positive DRB1*15:02 allele had a higher eGFR at diagnosis (124±23 versus 101±26 ml/min per 1.73 m2; P=0.004) and at the last follow-up (119±23 versus 101±26 ml/min per 1.73 m2; P=0.009) than patients without it.
Clinical Characteristics and Kidney Outcomes of Patients with M-Type Phospholipase A2 Receptor–Related Membranous Nephropathy with or without Histocompatibility Leukocyte Antigen Risk Alleles
In this study, the carrier rates of the four identified HLA risk alleles were not different between patients with PLA2R-related membranous nephropathy and healthy controls (Supplemental Table 2). The clinical characteristics of patients with PLA2R-related membranous nephropathy with or without any HLA risk alleles are shown in Table 1. During a median follow-up of 63 (IQR, 11–84) months, 65 (17%) patients developed a ≥40% eGFR decline. Of the 65 patients, 19 (29%) carried at least one of the four HLA risk alleles. Patients with any risk HLA alleles had a higher incidence of developing a ≥40% eGFR decline (41% versus 8%; P<0.001) than those without. The overall incidence rate of a ≥40% eGFR decline during follow-up is 4.2 cases per 100 person-years. The incidence rate in patients with any risk HLA alleles was 12.8 cases per 100 person-years, compared with 3.3 cases per 100 person-years in patients without any risk HLA alleles. Of the 65 patients who developed 40% eGFR decline, the median time to a 40% decline in eGFR was 4.52 (IQR, 0.73–6.8) years.
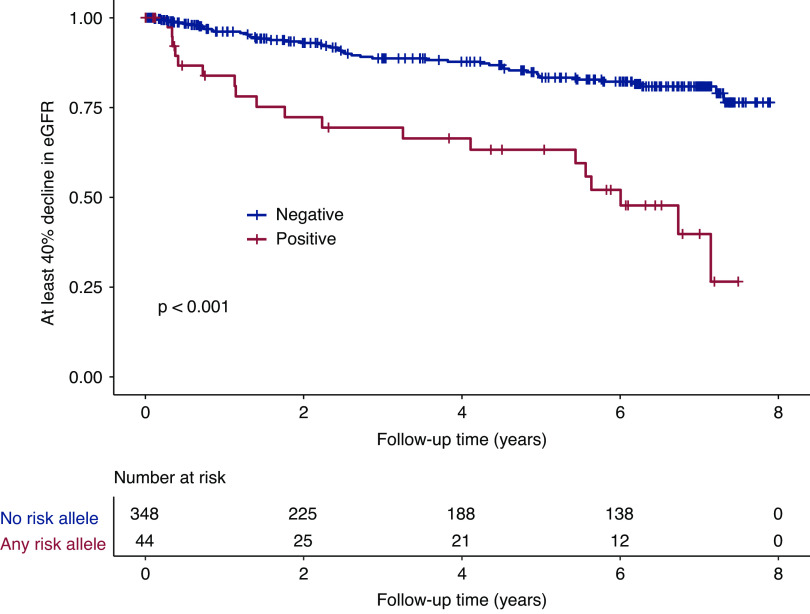
Kaplan–Meier analyses also demonstrated a significantly lower kidney survival for patients carrying any risk HLA alleles (Figure 1). At 7 years of follow-up, the cumulative kidney survival from a ≥40% decline in eGFR was 40% in patients with any risk HLA alleles (n=44), compared with 81% in patients without any risk HLA alleles (n=348) (Figure 1). The results were similar when the kidney outcome event was defined as a ≥50% decline in eGFR (Supplemental Figure 1). We also performed Kaplan–Meier analysis in the four identified risk HLA alleles separately, and the results are shown in Supplemental Figures 2–4. As shown in Table 2, after adjusting for age, sex, proteinuria, albumin, eGFR, and anti-PLA2R antibody levels, multivariable Cox analysis showed that carrying any of the four risk HLA alleles remained independently associated with a ≥40% decline in eGFR during follow-up (HR, 4.1; 95% CI, 2.3 to 7.1; P<0.001). The results were similar when we repeated the Cox analysis using a ≥50% eGFR decline as the kidney outcome (HR, 4.5; 95% CI, 2.4 to 8.3; P<0.001) (Supplemental Table 3).
Figure 1.
Kaplan–Meier analysis of kidney survival curves (≥40% decline in eGFR) in patients with M-type phospholipase A2 receptor–related membranous nephropathy. Compared with patients without any HLA risk allele possession (n=348), those with at least one of the four HLA risk alleles (n=44) had worse kidney outcomes during follow-up. HLA risk alleles associated with kidney outcome: DRB1*13:01, DQB1*06:03, DRB1*04:05, or DQB1*03:02.
Table 2.
Risk factors for assessing the risk of a ≥40% eGFR decline in patients with M-type phospholipase A2 receptor–related membranous nephropathy (n=392)
| Factor | Unadjusted Cox Regression | Multivariable Cox Regression | ||||
| Hazard Ratio | 95% Confidence Interval | P Value | Hazard Ratio | 95% Confidence Interval | P Value | |
| Any HLA risk alleles possessiona | 3.9 | 2.3 to 6.7 | <0.001 | 4.1 | 2.3 to 7.1 | <0.001 |
| Women | 0.42 | 0.23 to 0.76 | 0.004 | 0.59 | 0.31 to 1.11 | 0.10 |
| Age at baseline for every 10 yr older | 1.3 | 1.10 to 1.52 | 0.002 | 1.1 | 0.90 to 1.4 | 0.31 |
| Proteinuria at baseline, g/24 h, for every 1 g/24 h higher | 1.2 | 1.12 to 1.34 | <0.001 | 1.1 | 1.00 to 1.25 | 0.04 |
| Albumin at baseline, g/dl, for every 1 g/d lower | 0.51 | 0.31 to 0.83 | 0.007 | 0.88 | 0.50 to 1.55 | 0.65 |
| eGFR at baseline, for every 10 ml/min per 1.73 m2 higher | 0.81 | 0.74 to 0.88 | <0.001 | 0.88 | 0.78 to 1.00 | 0.06 |
| Anti-PLA2R antibodies, for every 10 U/ml higher | 1.01 | 1.01 to 1.02 | 0.001 | 1.01 | 1.00 to 1.02 | 0.02 |
PLA2R, M-type phospholipase A2 receptor.
HLA risk alleles: DRB1*13:01, DQB1*06:03, DRB1*04:05, or DQB1*03:02.
Relationship between the Susceptible Alleles and the Four Identified Progressive Risk Alleles
Both HLA DRB1*15:01 and DRB3*02:02 (or DRB1*03:01) were identified as susceptible HLA alleles in Chinese patients with idiopathic membranous nephropathy. In this study, among the 12 patients with positive DRB1*13:01-DQB1*06:03, all (100%) were positive for DRB3*02:02, and only four patients (33%) were positive for DRB1*15:01 (Supplemental Table 4). In patients with DRB1*15:01+DRB3*02:02−, none were positive for DRB1*13:01. As expected, patients with DRB1*13:01+DRB3*02:02+ had a significantly higher rate of kidney events than those with DRB1*13:01−DRB3*02:02+ (Supplemental Figure 2B). These data indicated that DRB1*13:01-DQB1*06:03 might interact with the membranous nephropathy–susceptible allele DRB3*02:02 but not with the susceptible allele DRB1*15:01 in disease progression. No specific distribution pattern could be observed between two other progressive risk alleles (DRB1*04:05 and DQB1*03:02) and the susceptible alleles.
Data from National Marrow Donor Program showed that people carrying DRB1*03:01 were most likely to be positive for DRB3*02:02, whereas those carrying DRB3*02:02 were positive for various DRB1 alleles, including DRB1*03:01 and DRB1*13:01 (Supplemental Figure 5). Remarkably, although all of the 12 patients with DRB1*13:01+ were positive for DRB3*02:02, none of them were positive for DRB1*03:01. Thus, the DRB3*02:02-DRB1*13:01 haplotype, but not the DRB3*02:02-DRB1*03:01 haplotype, was associated with poor kidney outcome in Chinese patients with PLA2R-related membranous nephropathy.
Discussion
The genetic background of idiopathic membranous nephropathy has been widely explored in the last decade, and the strongest association has been identified between HLA alleles and idiopathic membranous nephropathy. In patients originating from East Asia, two independent susceptible HLA haplotypes, DRB1*15:01 and DRB3*02:02 (or DRB1*03:01 linked with DRB3*02:02), are associated with the development of idiopathic membranous nephropathy (9,12). We previously found that almost all (99%) Chinese patients with PLA2R-related membranous nephropathy carry either or both DRB1*15:01 and DRB3*02:02 alleles (9). In this study, we further investigated whether HLA alleles were associated with kidney outcomes in patients with PLA2R-related membranous nephropathy. We found that four HLA alleles, DRB1*13:01, DQB1*06:03, DRB1*04:05, and DQB1*03:02, were associated with kidney outcomes independent of the clinical features. Among the four alleles, DRB1*13:01 and DQB1*06:03 were highly linked. These four HLA alleles were not associated with a greater susceptibility of idiopathic membranous nephropathy, but they could modify the disease phenotype and kidney outcome (22). Our results also showed that alleles associated with susceptibility to PLA2R-related membranous nephropathy (DRB3*02:02/DRB1*03:01 and DRB1*15:01) were not associated with adverse kidney function or outcome.
Interestingly, all patients with DRB1*13:01 were also positive for DRB3*02:02 (100%), but none of them were positive for DRB1*03:01 (0%). Among DRB1 alleles linked to DRB3*02:02, the DRB3*02:02-DRB1*03:01 haplotype is common (7%), whereas the DRB3*02:02-DRB1*13:01 haplotype (0.2%) is low in the Chinese population (23,24). The DRB1*03:01 allele was almost completely linked with DRB3*02:02, whereas the DRB3*02:02 allele was linked with many DRB1 alleles. Our results suggested that the DRB3*02:02-DRB1*13:01 haplotype, but not the DRB3*02:02-DRB1*03:01 haplotype, was independently associated with kidney prognosis in PLA2R-related membranous nephropathy.
The distribution of the four risk HLA alleles identified in this study differed greatly among populations. In the general Chinese population, DRB1*13:01 (2%) and DQB1*06:03 (2%) have low carrying rate, whereas DRB1*04:05 (12%) and DQB1*03:02 (12%) have common carrying rate (23–27). In European Whites, the carrying rates of DRB1*13:01 (11%), DQB1*06:03 (11%), and DQB1*03:02 (19%) are relatively common, whereas the carrying rate of DRB1*04:05 (1%) is at a low level. In the Caribbean Hispanic population, all four risk HLA alleles are relatively common, with DRB1*13:01 at 14%, DQB1*06:03 at 13%, DRB1*04:05 at 7%, and DQB1*03:02 at 19% (23). DRB1*13:01 is linked with the membranous nephropathy–susceptible allele DRB3*02:02 (linked with DQA1*0501), which is the strongest susceptible allele in European patients with membranous nephropathy. These data suggest that the DRB1*13:01 allele has a more important role in disease progression in Europeans than in Chinese individuals, whereas DRB1*04:05 has a more important role in Chinese individuals than Europeans.
Another study from China showed that DRB1*15:02 was associated with a lower eGFR at baseline and last follow-up and a higher percentage of kidney failure in unadjusted analysis, but the effect disappeared in multivariable analysis (20). Associations between DRB1*15:02 and kidney outcomes were not found in this study. It is not clear why the results of this study are inconsistent with the previous one. In fact, patients with the DRB1*15:02 allele were likely to have a more favorable kidney outcome. The inconsistent results could be due to different enrollment criteria between the two cohorts. The risk HLA alleles for kidney outcome could be different between patients with PLA2R-related membranous nephropathy and those with PLA2R-unrelated membranous nephropathy. The main goal of this study was to identify the HLA risk alleles associated with PLA2R-related membranous nephropathy; thus, patients with PLA2R-unrelated membranous nephropathy were not enrolled. All 392 patients were positive for both serum anti-PLA2R antibody (≥20 RU/ml) and glomerular PLA2R antigen staining. The previous study did not exclude patients with PLA2R-unrelated membranous nephropathy.
It is a limitation of this study that the separate signals of DRB1*04:05 and DQB1*03:02 did not meet the Bonferroni correction in the screening Cox analysis. However, on the basis of the following considerations, these two alleles were included for further analysis. First, the association signals of DRB1*04:05 and DQB1*03:02 in screening analysis were so strong that we cannot ignore them. Second, the Bonferroni correction is relatively conservative in this study because the 79 HLA alleles identified here were always linked with each other and were not independent signals. For example, the DRB1 locus is the most polymorphic locus of the human being, and there were 36 alleles identified from the DRB1 locus in this study (Supplemental Tables 1 and 2). If the alleles from DRB1 locus were considered as “independent” signals, these 36 alleles could be considered as “independent” signals for analysis. After filtering alleles with carrying rate ≤0.01, there could be only 22 alleles left. Third, after adjusting for DRB1*13:01/DQB1*06:03, the association signals of DRB1*04:05 and DQB1*03:02 were even reinforced, indicating DRB1*04:05 and DQB1*03:02 were strong independent risk alleles. We also considered the signals of DQA1*01:03 and DQA1*03:03. However, after adjusting for the three strongest risk alleles, the association signals of DQA1*01:03 (HR, 1.6; 95% CI, 0.69 to 3.8; P=0.26) and DQA1*03:03 (HR, 1.7; 95% CI, 0.23 to 12.3; P=0.60) disappeared. Fourth, on the basis of the immunologic pathogenesis of membranous nephropathy, it is reasonable to hypothesize that there could be multiple HLA alleles associated with kidney outcome. Thus, we combined these risk alleles together for further analysis and found that carrying any of the risk HLA alleles was significantly associated with kidney outcome (HR, 3.9; 95% CI, 2.3 to 6.7; P=5.9×10−7). After adjusting for clinical characteristics, the combined signal remained significant. Even if the Bonferroni correction was applied, the results of the combined alleles reached the statistical significant level. Thus, we report the combined analysis as the main results in this study.
Taken together, our results showed that DRB1*13:01/DQB1*06:03, DRB1*04:05, and DQB1*03:02 were independently associated with poor kidney prognosis in PLA2R-related membranous nephropathy. Patients with any of the risk alleles had a remarkable adjusted HR of 4.1 (95% CI, 2.3 to 7.1) for a ≥40% eGFR decline compared with those without. The distribution of the four risk HLA alleles differs greatly among populations. These findings need to be validated in other membranous nephropathy cohorts and tested in populations with diverse ancestries, including those with a higher frequency of DRB1*13:01, to test if differences in these alleles explain some of the differences in prognosis of membranous nephropathy between populations.
Disclosures
All authors have nothing to disclose.
Funding
This work is supported by Natural Science Foundation of China grants 81970620 and 81500547, Natural Science Foundation of Jiangsu Province grant BK20150560, and Young Medical Talents Program of Jiangsu Province grant QNRC2016894.
Supplementary Material
Acknowledgments
We thank the physicians, research nurses, patients, and healthy volunteers who contributed to this study.
Data Sharing Statement
The datasets used and analyzed during this study are available from the corresponding author on reasonable request.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.18021120/-/DCSupplemental.
Supplemental Table 1. List of hazard ratios of HLA alleles in Cox proportional hazards models to assess associations between HLA alleles and kidney outcome event (≥40% eGFR decline during follow-up) in patients with PLA2R-related membranous nephropathy.
Supplemental Table 2. The carrying rate of HLA alleles in patients and healthy controls in this study.
Supplemental Table 3. Risk factors for assessing risk of ≥50% eGFR decline in patients with PLA2R2-related membranous nephropathy (n = 392).
Supplemental Table 4. Relationship between the susceptible alleles and the four identified progressive risk alleles.
Supplemental Figure 1. Kaplan–Meier analysis of kidney survival curves (≥50% decline in eGFR) in patients with PLA2R-related membranous nephropathy.
Supplemental Figure 2. Kaplan–Meier analysis of kidney survival curves (≥40% decline in eGFR) in patients with DRB1*13:01+, DRB1*13:01-, DRB1*13:01+DRB3*02:02+, and DRB1*13:01-DRB3*02:02+.
Supplemental Figure 3. Kaplan–Meier analysis of kidney survival curves (≥40% decline in eGFR) in patients with HLA DRB1*04:05+.
Supplemental Figure 4. Kaplan–Meier analysis of kidney survival curves (≥40% decline in eGFR) in patients with HLA DQB1*03:02+.
Supplemental Figure 5. Linkage map of HLA-DRB3/4/5 with HLA-DRB1 in the National Marrow Donor Program of the Chinese population.
References
- 1.Hou JH, Zhu HX, Zhou ML, Le WB, Zeng CH, Liang SS, Xu F, Liang DD, Shao SJ, Liu Y, Liu ZH: Changes in the spectrum of kidney diseases: An analysis of 40,759 biopsy-proven cases from 2003 to 2014 in China. Kidney Dis 4: 10–19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, Wang G, Chen N, Lu T, Nie S, Xu G, Zhang P, Luo Y, Wang Y, Wang X, Schwartz J, Geng J, Hou FF: Long-term exposure to air pollution and increased risk of membranous nephropathy in China. J Am Soc Nephrol 27: 3739–3746, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck LH Jr., Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, Klein JB, Salant DJ: M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med 361: 11–21, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronco P, Debiec H: Molecular pathogenesis of membranous nephropathy. Annu Rev Pathol 15: 287–313, 2020 [DOI] [PubMed] [Google Scholar]
- 5.Qin HZ, Zhang MC, Le WB, Ren Q, Chen DC, Zeng CH, Liu L, Zuo K, Xu F, Liu ZH: Combined assessment of phospholipase A2 receptor autoantibodies and glomerular deposits in membranous nephropathy. J Am Soc Nephrol 27: 3195–3203, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin W, Beck LH Jr., Zeng C, Chen Z, Li S, Zuo K, Salant DJ, Liu Z: Anti-phospholipase A2 receptor antibody in membranous nephropathy. J Am Soc Nephrol 22: 1137–1143, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomas NM, Beck LH Jr., Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, Dolla G, Hoxha E, Helmchen U, Dabert-Gay AS, Debayle D, Merchant M, Klein J, Salant DJ, Stahl RAK, Lambeau G: Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med 371: 2277–2287, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi S, Debiec H, Madden B, Charlesworth MC, Morelle J, Gross L, Ravindran A, Buob D, Jadoul M, Fervenza FC, Ronco P: Neural epidermal growth factor-like 1 protein (NELL-1) associated membranous nephropathy. Kidney Int 97: 163–174, 2020 [DOI] [PubMed] [Google Scholar]
- 9.Le WB, Shi JS, Zhang T, Liu L, Qin HZ, Liang S, Zhang YW, Zheng CX, Jiang S, Qin WS, Zhang HT, Liu ZH: HLA-DRB1*15:01 and HLA-DRB3*02:02 in PLA2R-related membranous nephropathy. J Am Soc Nephrol 28: 1642–1650, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, Stephens HA, Laundy V, Padmanabhan S, Zawadzka A, Hofstra JM, Coenen MJ, den Heijer M, Kiemeney LA, Bacq-Daian D, Stengel B, Powis SH, Brenchley P, Feehally J, Rees AJ, Debiec H, Wetzels JF, Ronco P, Mathieson PW, Kleta R: Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 364: 616–626, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Xie J, Liu L, Mladkova N, Li Y, Ren H, Wang W, Cui Z, Lin L, Hu X, Yu X, Xu J, Liu G, Caliskan Y, Sidore C, Balderes O, Rosen RJ, Bodria M, Zanoni F, Zhang JY, Krithivasan P, Mehl K, Marasa M, Khan A, Ozay F, Canetta PA, Bomback AS, Appel GB, Sanna-Cherchi S, Sampson MG, Mariani LH, Perkowska-Ptasinska A, Durlik M, Mucha K, Moszczuk B, Foroncewicz B, Pączek L, Habura I, Ars E, Ballarin J, Mani LY, Vogt B, Ozturk S, Yildiz A, Seyahi N, Arikan H, Koc M, Basturk T, Karahan G, Akgul SU, Sever MS, Zhang D, Santoro D, Bonomini M, Londrino F, Gesualdo L, Reiterova J, Tesar V, Izzi C, Savoldi S, Spotti D, Marcantoni C, Messa P, Galliani M, Roccatello D, Granata S, Zaza G, Lugani F, Ghiggeri G, Pisani I, Allegri L, Sprangers B, Park JH, Cho B, Kim YS, Kim DK, Suzuki H, Amoroso A, Cattran DC, Fervenza FC, Pani A, Hamilton P, Harris S, Gupta S, Cheshire C, Dufek S, Issler N, Pepper RJ, Connolly J, Powis S, Bockenhauer D, Stanescu HC, Ashman N, Loos RJF, Kenny EE, Wuttke M, Eckardt KU, Köttgen A, Hofstra JM, Coenen MJH, Kiemeney LA, Akilesh S, Kretzler M, Beck LH, Stengel B, Debiec H, Ronco P, Wetzels JFM, Zoledziewska M, Cucca F, Ionita-Laza I, Lee H, Hoxha E, Stahl RAK, Brenchley P, Scolari F, Zhao MH, Gharavi AG, Kleta R, Chen N, Kiryluk K: The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat Commun 11: 1600, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Z, Xie LJ, Chen FJ, Pei ZY, Zhang LJ, Qu Z, Huang J, Gu QH, Zhang YM, Wang X, Wang F, Meng LQ, Liu G, Zhou XJ, Zhu L, Lv JC, Liu F, Zhang H, Liao YH, Lai LH, Ronco P, Zhao MH: MHC class II risk alleles and amino acid residues in idiopathic membranous nephropathy. J Am Soc Nephrol 28: 1651–1664, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekula P, Li Y, Stanescu HC, Wuttke M, Ekici AB, Bockenhauer D, Walz G, Powis SH, Kielstein JT, Brenchley P, Eckardt KU, Kronenberg F, Kleta R, Köttgen A; GCKD Investigators: Genetic risk variants for membranous nephropathy: Extension of and association with other chronic kidney disease aetiologies. Nephrol Dial Transplant 32: 325–332, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiri M, Honda K, Kashiwase K, Mabuchi A, Suzuki H, Watanabe K, Nakayama M, Watanabe T, Doi K, Tokunaga K, Noiri E: High-density association mapping and interaction analysis of PLA2R1 and HLA regions with idiopathic membranous nephropathy in Japanese. Sci Rep 6: 38189, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullich G, Ballarín J, Oliver A, Ayasreh N, Silva I, Santín S, Díaz-Encarnación MM, Torra R, Ars E: HLA-DQA1 and PLA2R1 polymorphisms and risk of idiopathic membranous nephropathy. Clin J Am Soc Nephrol 9: 335–343, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeed M, Beggs ML, Walker PD, Larsen CP: PLA2R-associated membranous glomerulopathy is modulated by common variants in PLA2R1 and HLA-DQA1 genes. Genes Immun 15: 556–561, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Zhang XD, Cui Z, Zhao MH: The genetic and environmental factors of primary membranous nephropathy: An overview from China. Kidney Dis 4: 65–73, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kidney Disease: Improving Global Outcomes (KDIGO) : KDIGO Clinical Practice Guideline for Glomerulonephritis. Available at: https://kdigo.org/wp-content/uploads/2017/02/KDIGO-2012-GN-Guideline-English.pdf. Accessed November 16, 2020 [Google Scholar]
- 19.De Vriese AS, Glassock RJ, Nath KA, Sethi S, Fervenza FC: A proposal for a serology-based approach to membranous nephropathy. J Am Soc Nephrol 28: 421–430, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HY, Cui Z, Xie LJ, Zhang LJ, Pei ZY, Chen FJ, Qu Z, Huang J, Zhang YM, Wang X, Wang F, Meng LQ, Cheng XY, Liu G, Zhou XJ, Zhang H, Debiec H, Ronco P, Zhao MH: HLA class II alleles differing by a single amino acid associate with clinical phenotype and outcome in patients with primary membranous nephropathy. Kidney Int 94: 974–982, 2018 [DOI] [PubMed] [Google Scholar]
- 21.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration): A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck LH Jr., Salant DJ: Deep pockets are not necessarily a good thing in membranous nephropathy: Evidence for a modifier allele. Kidney Int 94: 855–857, 2018 [DOI] [PubMed] [Google Scholar]
- 23.Gragert L, Madbouly A, Freeman J, Maiers M: Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 74: 1313–1320, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Galarza FF, McCabe A, Santos EJMD, Jones J, Takeshita L, Ortega-Rivera ND, Cid-Pavon GMD, Ramsbottom K, Ghattaoraya G, Alfirevic A, Middleton D, Jones AR: Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res 48[D1]: D783–D788, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin Qin P, Su F, Xiao Yan W, Xing Z, Meng P, Chengya W, Jie S: Distribution of human leucocyte antigen-A, -B and -DR alleles and haplotypes at high resolution in the population from Jiangsu province of China. Int J Immunogenet 38: 475–481, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Kwok J, Guo M, Yang W, Lee CK, Ho J, Tang WH, Chan YS, Middleton D, Lu LW, Chan GC: HLA-A, -B, -C, and -DRB1 genotyping and haplotype frequencies for a Hong Kong Chinese population of 7595 individuals. Hum Immunol 77: 1111–1112, 2016 [DOI] [PubMed] [Google Scholar]
- 27.Kwok J, Guo M, Yang W, Lee CK, Chan NK, Ho J, Tang WH, Chan YS, Middleton D, Lu LW, Chan GC: HLA-A, -B and -DRB1 genotyping and haplotype frequencies of 3892 cord blood units in the Hong Kong Chinese Cord Blood Registry. Hum Immunol 77: 1109–1110, 2016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.