Figure 2.
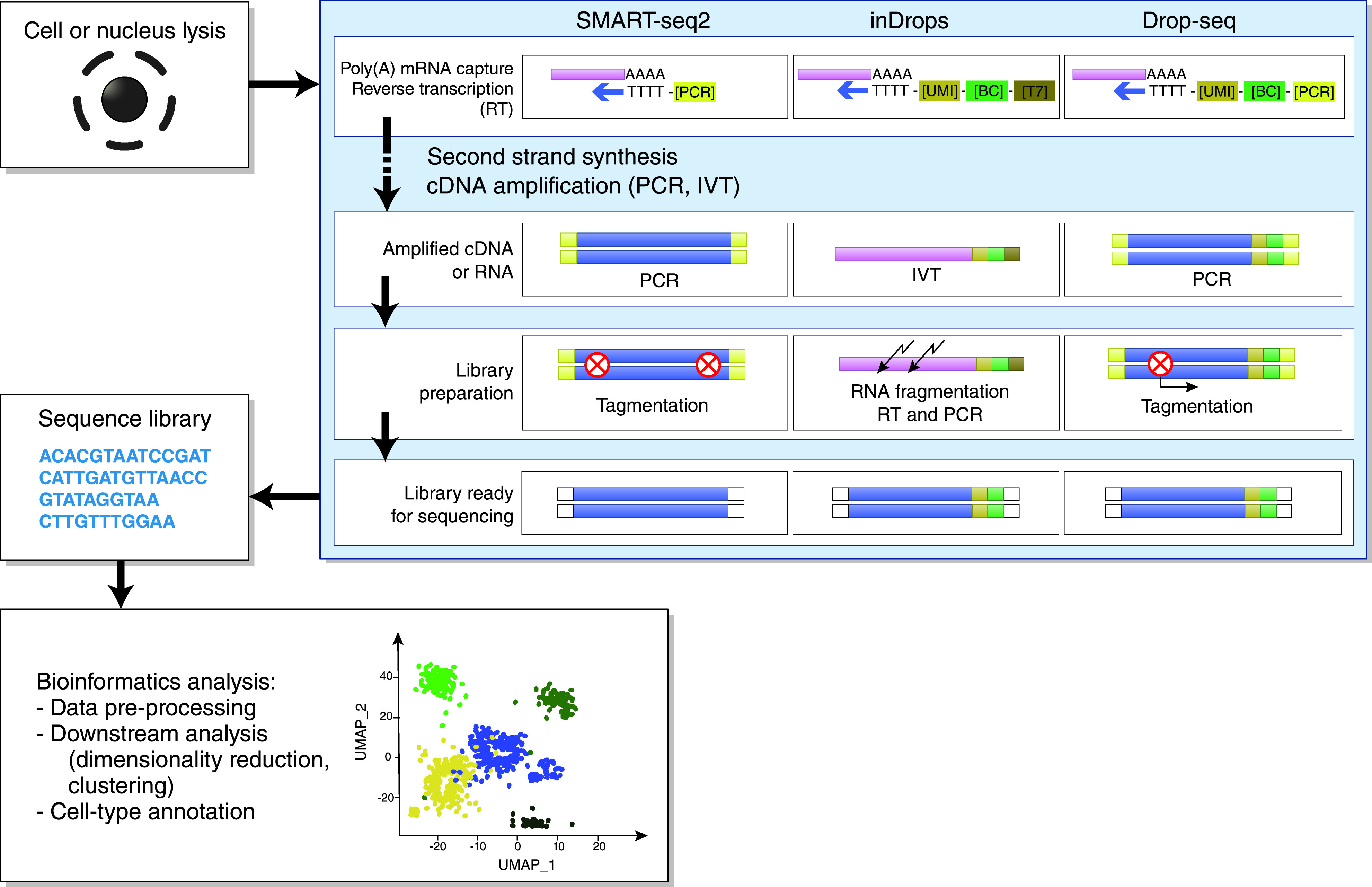
General workflow of scRNA-seq and snRNA-seq experiments, continued. Individual cells or nuclei are lysed and transcripts from single cells or single nuclei are captured using poly[T] oligonucleotide primers that hybridize with the polyadenylated (poly[A]) mRNA.3,82 These primers may also contain other oligonucleotide sequences, including a cell barcode (BC), unique molecular identifier (UMI), and adaptor sequence for PCR or RNA polymerase promotor (T7 promotor) for in vitro transcription (IVT).3 Next, a reverse transcription (RT) reaction synthesizes the first and second strand of cDNA, which is subsequently amplified by either PCR or IVT. Amplified cDNA molecules are fragmented (using enzymatic tagmentation or chemical fragmentation) and ligated with adaptors for sequencing.3 A sequence library is created and, finally, using advanced bioinformatics analysis, sequencing data are translated into cellular expression patterns, leading to the identification of distinct cell clusters.83 Adapted from refs. 3,4,81,82. UMAP, Uniform Manifold Approximation and Projection.
