Figure 6.
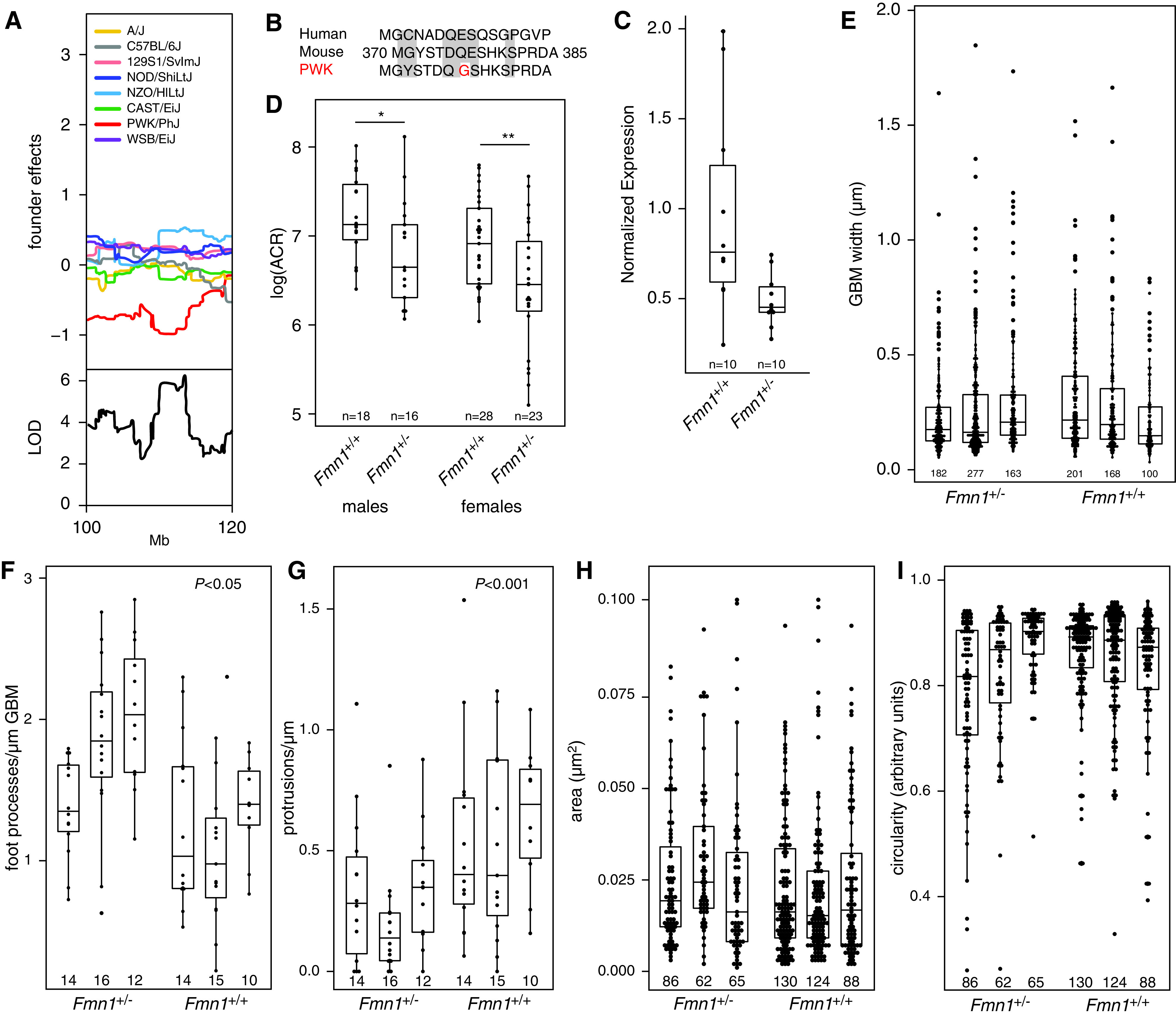
Identification and characterization of Fmn1 as a candidate gene. (A) Allele effect plot of the Chr 2 locus shows that animals that inherited the allele from the PWK founder strain have lower ACR. (B) Analysis of the founder genomes in the 95% confidence interval of the Chr 2 locus shows that the PWK genome has a genetic variant in the coding region of Fmn1 that leads to an amino acid change unique to PWK compared with the seven other founder strains of the DO-XLAS cohort and humans. (C) Quantitative PCR shows that Fmn1+/− mice have an approximately 50% lower Fmn1 expression compared with wild-type mice. (D) Col4a5 mutant mice with only one functional Fmn1 allele (Fmn1+/−) have lower ACR in both males and females compared with Col4a5 mutant mice with two functional Fmn1 alleles (Fmn1+/+). Quantification in male mice using transmission electron microscopy does not show differences in (E) GBM width, (H) protrusion area, or (I) protrusion circularity, but there are significant differences in (F) the number of FPs per micrometer of GBM and (G) the number of protrusions per micrometer of GBM. The number under each bar indicates the number of measurements in each animal. LOD, logarithm of the odds. *P=0.05; **P=0.01.
