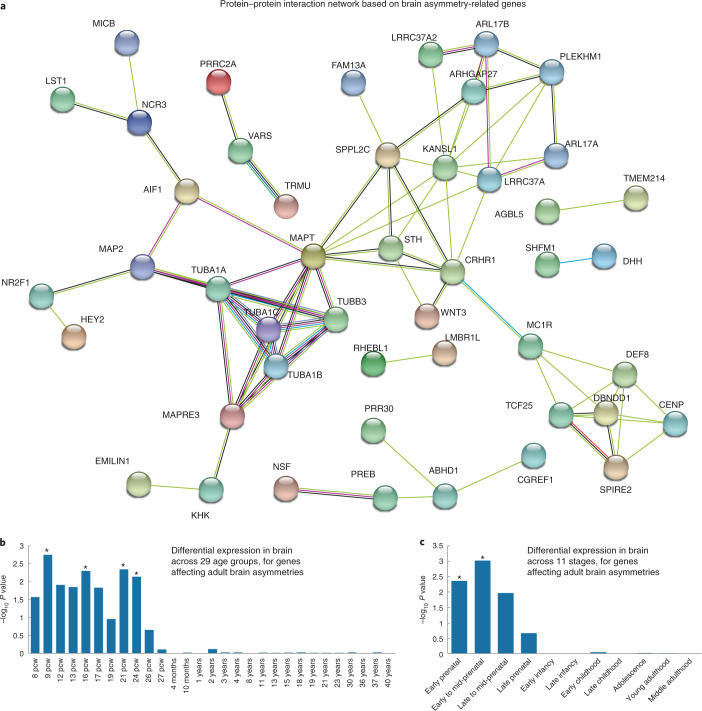
Fig. 4. Functional annotations of variants associated with brain asymmetry.
a, Brain asymmetry-associated genes integrated into a protein–protein interaction network. Proteins are represented by nodes. Edges between nodes represent different types of protein–protein interactions according to the STRING database (Methods), including known interactions (turquoise and dark purple represent interactions identified by curated databases and biological experiments, respectively), predicted interactions (green, red and blue represent interactions predicted by gene neighbourhood, gene fusions and gene co-occurrence, respectively) and others (yellow, black and light purple represent interactions determined by text mining, co-expression and protein homology, respectively). Coloured nodes represent the queried proteins. Only medium-confidence (>0.4) links were retained, and disconnected proteins are not shown. b, Relation between gene-based association with brain asymmetries and relatively higher mRNA expression in the human brain at particular ages, using BrainSpan data from 29 age groups. Asterisks indicate significant age groups meeting P < 0.05 with FDR correction. pcw, post-conceptional weeks. c, Relation between gene-based association with brain asymmetries and relatively higher mRNA expression in the human brain at particular ages, using BrainSpan data from 11 defined age groups. Asterisks indicate significant groups meeting P < 0.05 with FDR correction.