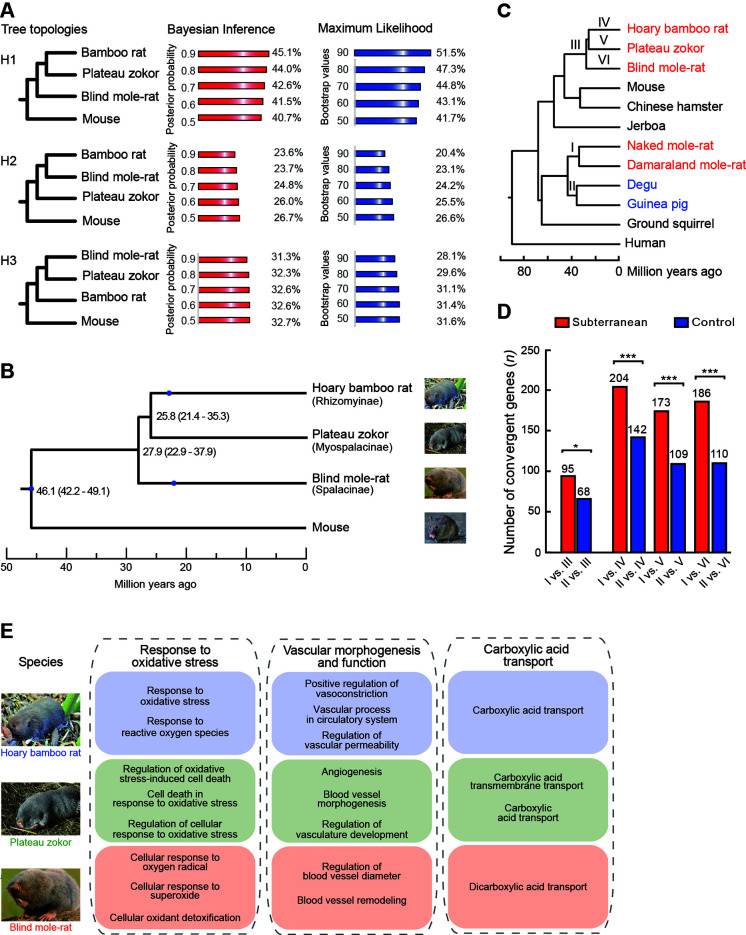
Figure 1.
Reconstruction of phylogeny among spalacids and genome-wide convergent analyses
A: Proportions of individual genes supporting each possible phylogenetic topology based on Bayesian inference and maximum-likelihood approaches are listed behind different supporting cutoffs. B: Estimated divergence times among Myospalacinae, Spalacinae, and Rhizomyinae. Blue spots indicate fossil constraints for calibration of estimated divergence times. Numbers at nodes are molecular dates estimated after correction; values in parentheses are 95% credibility intervals. C: Phylogeny with divergence times was used to detect molecular convergences. I, II, III, IV, V, and VI denote branches where molecular convergences are detected. D: Number of convergent genes is significantly larger for subterranean branch pairs than for their respective control branch pairs at the genomic scale. *: P<0.05;***: P<0.001. E: Functional enrichment analyses of convergent genes between subterranean African mole-rats and spalacids.